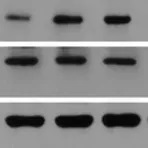
Enzymes, like Horseradish Peroxidase (HRP) or Alkaline Phosphatase (AP), can conjugate to an antibody via covalent linkages. Enzyme conjugated antibodies are used in conjunction with a substrate, either fluorogenic, colorimetric, or chemiluminescent, to create a detectable signal. The non-detectable, soluble substrate is converted by the enzyme into a detectable (and normally insoluble) form. Enzyme conjugated antibodies enable signal amplification; as the enzyme is able to convert many molecules of the substrate, the length of time the reaction is allowed to continue determines the strength of the signal.
Horseradish Peroxidase
Horseradish peroxidase (HRP) is an enzyme derived from the root of Armoracia rusticana (a.k.a. the horseradish plant) that is commonly used as a reporter protein. HRP conjugated secondary antibodies can either be used for colorimetric detection, where the HRP catalyses the conversion of a chromogenic substrate to a coloured precipitate, or chemiluminescent detection, where the HRP produces a signal by oxidising a chemiluminescent substrate to a form which emits light.
Multiple HRP molecules can bind to a single secondary antibody, as such, HRP conjugated secondary antibodies can be utilised for signal enhancement and enable detection of proteins expressed at low levels. HRP also has a high turnover rate which enables HRP conjugated secondary antibodies to generate a strong signal in a short amount of time (often within five minutes).
Alkaline Phosphatase
Alkaline phosphatase (AP) conjugated secondary antibodies are commonly used for ELISA and western blotting. They are also sometimes used for immunohistochemistry, however, their large size may limit penetration into tissues. It is worth noting, that whilst HRP generates a maximum signal quickly, often within five minutes, the signal from AP gradually increases and peaks at around an hour. AP generates a very stable signal which can last several days; this is particularly useful if multiple exposures are needed, especially over a matter of days.
Trying to decide between HRP and AP for your blot?
Use our table below to help with the decision.
| AP | HRP |
|---|
| Sensitive | Sensitive |
| Long lasting signal (24-48 hours) | Quick max signal (~5 minutes) |
| Useful for multiple exposures | Inhibited by azides |
| Inhibited by phosphate buffers | Economical |
Biotin
Biotin is a small, water-soluble B vitamin that can be bound by avidin, NeutrAvidin, and streptavidin with high affinity. Due to its small size, binding with biotin does not typically affect a protein’s biological activity. Multiple biotin molecules can conjugate to a single secondary antibody, as such, biotin conjugated secondary antibodies can be utilised for signal enhancement and enable detection of proteins expressed at low levels. Visualisation happens through biotin / streptavidin interaction whereby the streptavidin is bound to either HRP or a fluorescent probe. By utilising a biotin conjugated secondary antibody, the same secondary antibody can be used in multiple applications simply by switching the streptavidin used.