HOW TO MAXIMIZE YEAST TRANSFORMATION EFFICIENCY
TIPS AND BEST PRACTICES FOR TRANSFORMING POPULAR YEAST STRAINS
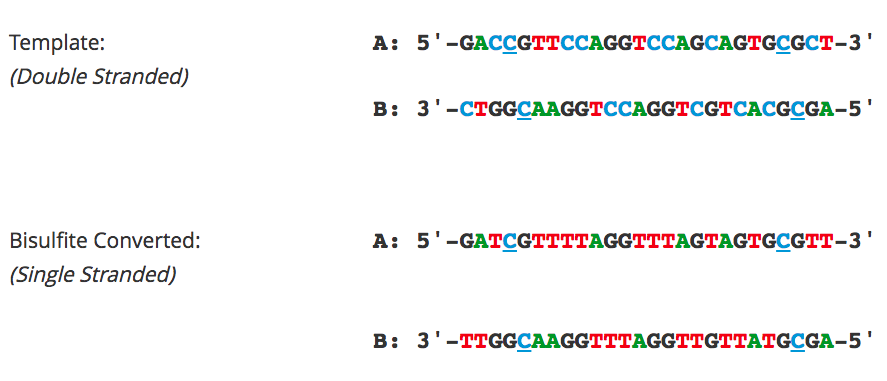
Introducing foreign DNA to yeast cells is essential for two-hybrid system screening and many other genetic manipulation techniques. The transformation efficiency of yeast cells is typically magnitudes lower than that of Escherichia coli due to the added challenges associated with permeating the fungal cell wall. However, implementing the following tips can help maximize the transformation efficiency of yeast species such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, Pichia pastoris, and Candida albicans.
- Cell Growth State: Use cells in the mid-log phase to produce the most transformants. Early or late log-phase cells yield comparatively fewer transformants.
- Cell Density: The optimal cell density for transformation is between 5 x 106 and 2 x 107 cells/ml (0.8-1.0 OD600). Yeast cultures with cell densities at the high end of this range typically report the highest transformation efficiencies.
- Heat Shocking: Because of their hardy cell walls, yeast cells must be heat-shocked more intensively than E. coli. To ensure high transformation efficiencies, yeast should be heat-shocked for 45 minutes.
- Plating Media: Not all commercially available media are created equal. Our results show that Difco media are the most reliable for maximizing transformation efficiency.
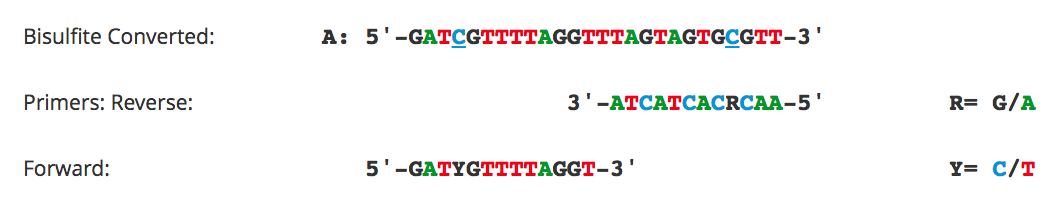
- DNA Input: For circular DNA such as plasmids, the transformation efficiency stops increasing linearly for DNA inputs above 1 µg. For integrative transformation with linearized DNA, higher inputs of up to 5 µg of DNA are recommended. The DNA extraction method is crucial, as highly pure DNA should be used for transformation.
Transformation efficiency is also species-dependent, as some strains are more susceptible to transformation than others. The following table provides a range of transformation efficiencies to expect from four prominent yeast strains using traditional transformation protocols.
Strain | Transformation Efficiency (cfu/µg DNA) |
|---|---|
S. cerevisiae | 104 – 106 |
S. pombe | 103 – 105 |
P. pastoris | 103 – 106 |
C. albicans | 102 – 104 |
It is important to note that the true transformation efficiency can vary significantly depending on the transformation method used, quality of the DNA, and other experimental parameters. For applications such as two-hybrid system screening or library screening that require high transformation efficiencies, it is imperative that researchers select a reliable transformation method such as the Frozen-EZ Yeast Transformation II Kit for optimal results. As an industry leader in yeast products, Zymo Research is dedicated to helping scientists streamline and elevate their transformations with innovative technologies.
REFERENCES
- https://zymoresearch.eu/blogs/blog/how-to-maximize-yeast-transformation-efficiency
February 07, 2024