04/30
2020
Lateral Flow Readout for CRISPR/Cas-based detection strategies
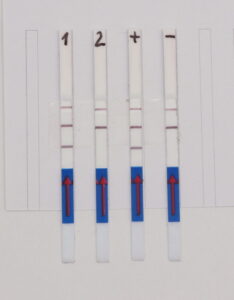
Test– and Control-line of the HybriDetect

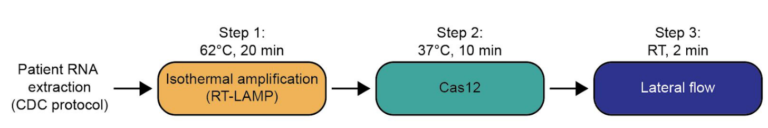
SHERLOCK and DETECTR are combinations of isothermal amplification and sensitive CRISPR/Cas-based amplicon recognition. The compatibility with the rapid, simple and equipment-free HybriDetect lateral flow platform is an essential feature of these methods. Interpretation of the lateral flow strips is very intuitive and easy.
The way HybriDetect is used for CRISPR applications can lead to confusions about the Test- and the Control-line. The following figure explains this difference.
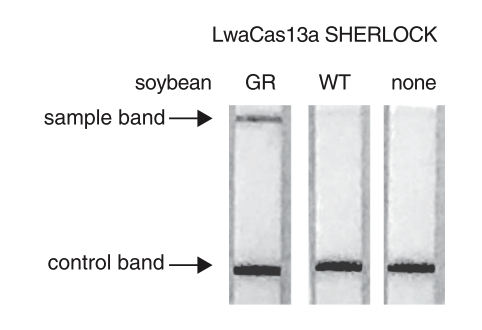
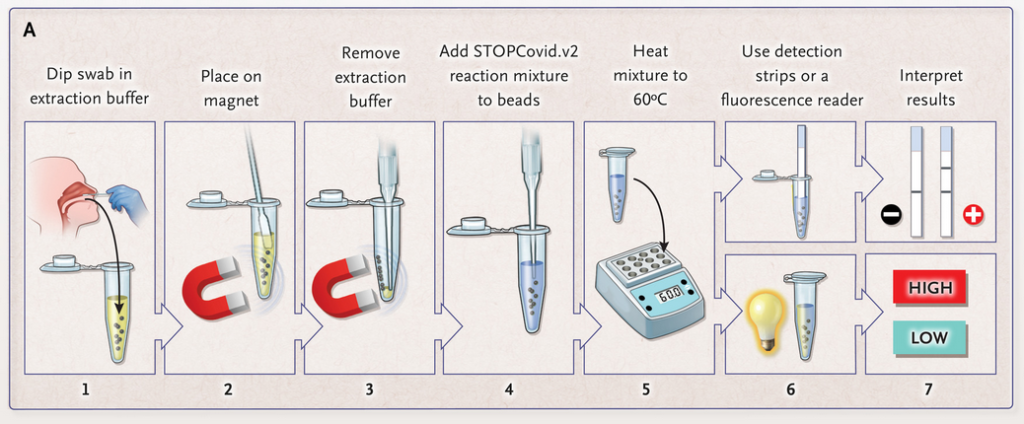
As shown in Figure 1, the interpretation of the test strips can vary using different detection strategies. Compared to the HybriDetect instructions for use, the T- and C-lines are swapped in CRSIPR/Cas-associated detection strategies. This leads to a completely different assessment of the resulting signals. The following article is intended to explain this discrepancy in detail and it provides some important information on how to use the HybriDetect in combination with the SHERLOCK / DETECTR method.
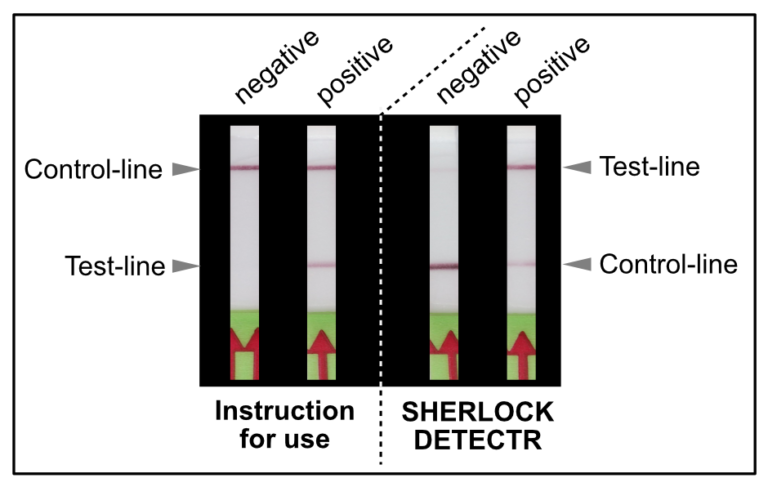
Figure 1. Difference of test result interpretation between the HybriDetect Instructions for use and the SHERLOCK-/DETECTR-protocol guidelines
What defines the control- and test-lines?
The “T and the “C” in Lateral Flow Devices (LFDs)
The presence of at least one visually detectable test-line (T-line) is the essential feature of LFDs. In most cases the presence of the T-line correlates with a positive test result. Additionally, a control-line (C-line) confirms the general functionality of LFA. This is absolutely necessary for differentiation between valid negative results and invalid, non-interpretable test results.
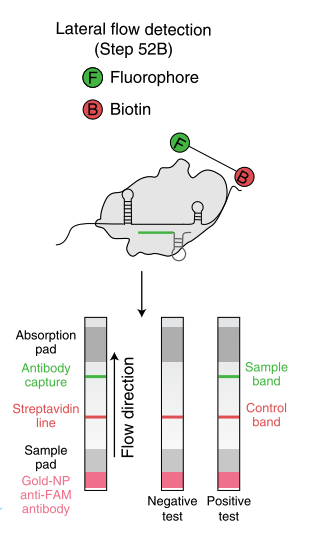
The HybriDetect design is basically comparable to the structure of a classic LFD, but the idea behind this detection platform differs.
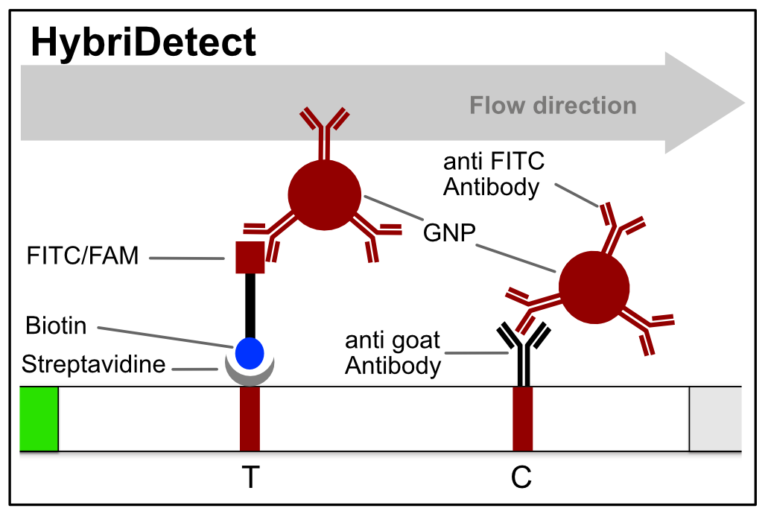
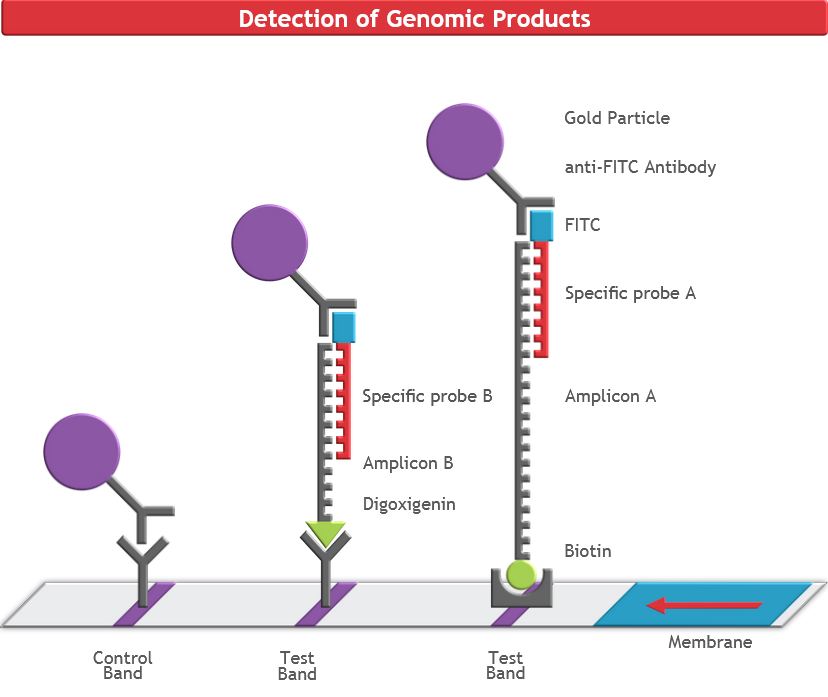
Figure 2. Composition of the HybriDetect. Biotin and FITC/FAM are introduced into LFA through sample application. The control-line (C) will show up, if you run the HybriDetect without sample. GNP – Goldnanoparticle
HybriDetect and signal generation strategies
The idea behind the HybriDetect platform differs fundamentally from “classic” LFAs. The HybriDetect and HybriDetect 2T are universal test strips, developed to work in multiple detection concepts and not in just one setting. They can be used to detect amplicons, proteins, ribosomal RNAs, whole cells or metabolites. Over the years various detection strategies have been developed, in which the HybriDetect is used.
In most constellations, biotin and FITC are somehow incorporated into analytes or reaction products, which results in the formation of the predefined test-line(s). Popular applications combined with HybriDetect readout are especially DNA amplification techniques such as PCR, RPA or LAMP. However, these LFA-compatible methods have one feature in common: they are designed according to the described Label Incorporation Strategy.
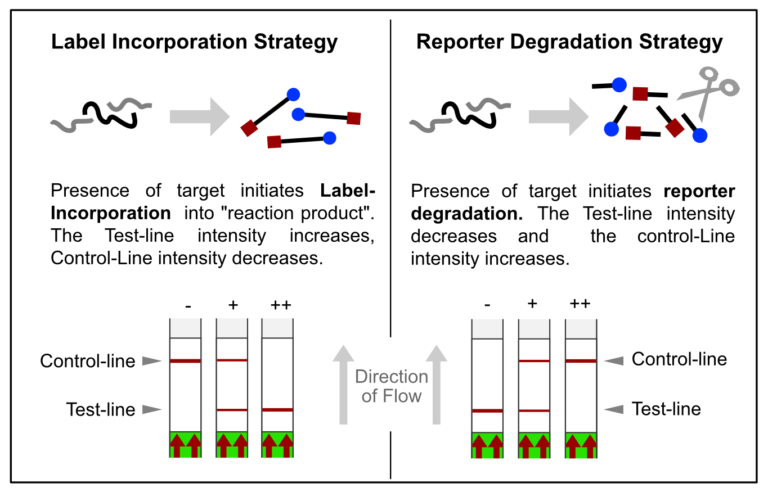
Figure 3. Overview of two LFA-related signal generation stratefies. Label incorporation strategy leads to Test-line formation, whereas reporter degradation will end up in decreased T-line and increased C-line intensity.
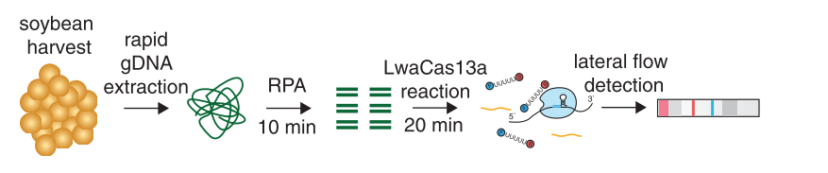
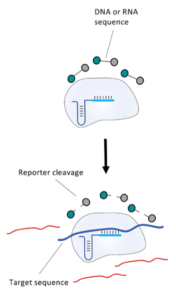
In contrast to this, methods such as SHERLOCK or DETECTR generate amplification products, which do not directly contribute to signal generation. Rather, CRISPR complexes are able to specifically and sensitively recognize defined amplicon-related sequences. Successful recognition leads to the activation of the Cas-protein associated collateral nuclease activity. The ssRNA- / ssDNA- / dsDNA – degrading activity is the basis for signal generation in LFA. This is achieved by using the reporter degradation strategy.
The inverse approach – degradation of the “original” T-line
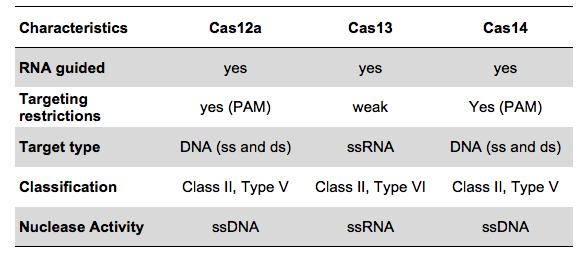
The key to the functionality of a CRISPR-associated nuclease assay combined with a lateral flow readout is the use of a so-called reporter. Such reporters are short single-stranded DNA- or RNA-Oligos with terminal biotin- and FITC-/ FAM-modifications. Reporter sequences are chosen according to cleavage preferences of the used Cas-protein. The following table shows a brief overview of some reporters, which are used in the most common CRISPR/Cas-based detection systems.
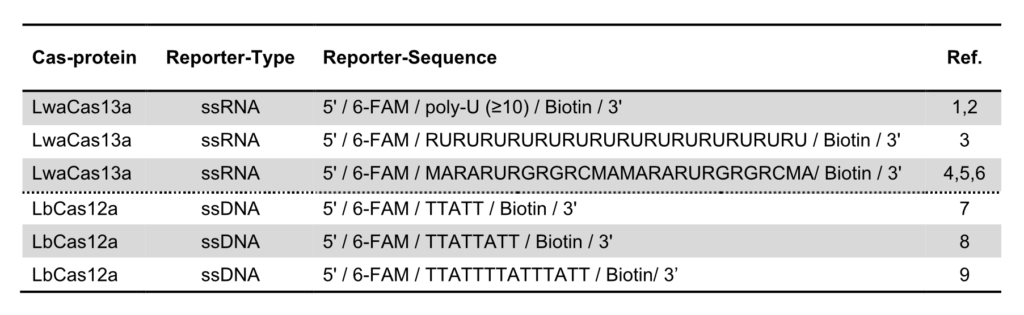
Table 1. Overview of reporter sequences used in CRISPR/Cas-based detection methods combined with Lateral Flow readout
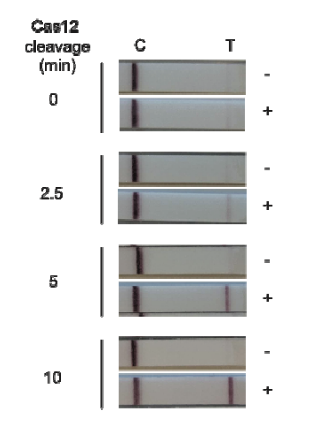
An intense test-line appears if a defined amount of reporter is analyzed with the HybriDetect. But if reporter degradation is initiated due to the onset of nuclease activity, the intensity of the T-line consequently decreases.
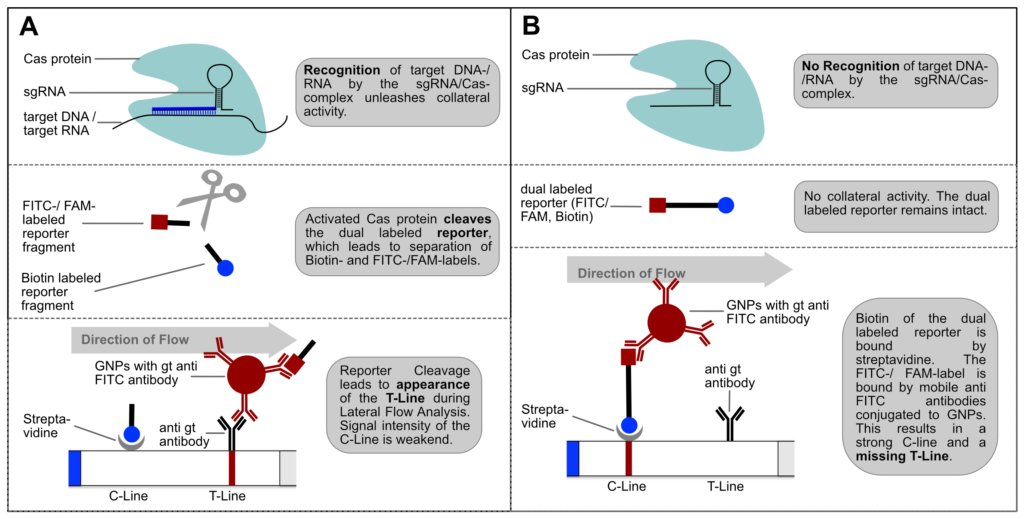
If the reporter is used in a very defined concentration, the majority of the gold conjugate is trapped at the T-line, which leads to an almost complete extinction of the C-line. It is precisely this effect, that enables the T- and C-line swap for CRISPR/Cas-associated detection methods. Ultimately, it’s just a matter of definition and there is no change in HybriDetect composition! The following figure explains the CRISPR/Cas-based detection strategy with newly defined T- and C-line.
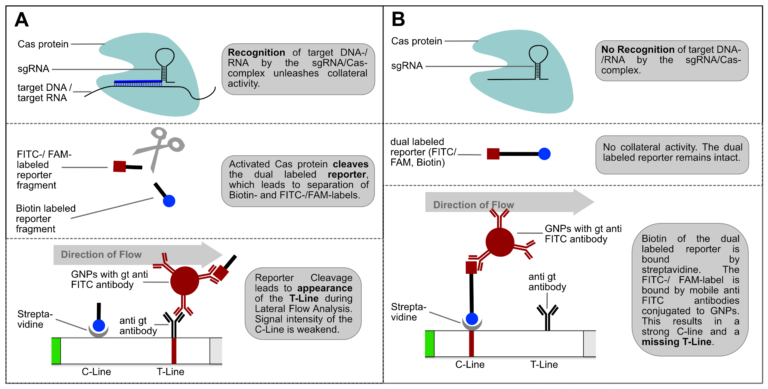
Figure 4. Switching C- and T-line: General mechanism of CRISPR/Cas-mediated detection of nucleic acids via HybriDetect Lateral Flow. (A) The presence of the genetic target leads to a positive test result. (B) The absence of the specific genetic target leads to a negative test result.
Switching test- and control-line has two major advantages: An increasing or appearing signal correlates with a positive test result. Negative results are interpreted if no signal appears. This positive correlation is much more logical for most people and avoids intuitive misinterpretation. This is a very important point when it comes to a point-of-care-compatible, simple and intuitive test interpretation. But it’s not only advantageous from a psychological perspective. Increasing intensities are easier to interpret for the human eye, which is a performance related argument for a more sensitive analysis.
For these reasons, the following part of the article swaps the C and T lines according to the guidelines of current papers on SHERLOCK- and DETECTR-methods.
BTW: Determining nuclease activity with lateral flow and a reporter is not entirely new. This form of lateral flow-based readout was already used in 2015 in connection with prostate cancer diagnosis and correlating DNAse I activity. A dsDNA reporter is used here in combination with a selfmade teststrip. This LFD has only one test-line, which is analogous to the control-Line of the HybriDetect. A loss of signal correlated with an increasing DNase I activity. (10)
How to use the reporter to eliminate the Test-line?
The almost complete extinction of the T-line is the basis for an easy and intuitive interpretation of the test strips. In order to eliminate T-line intensity as much as possible, it is necessary to determine the required amount of reporter in the LFA. The goal is to find the perfect amount of reporter, that the majority of the gold conjugate is retained at the C-Line. Only few gold nanoparticles will be able to travel to the T-line, leading to a missing or very weak T-line. For an almost complete elimination of the T-Line a special amount of reporter is needed.

Figure 5. Impact of reporter amount on C- and T-line intensities. A relatively long ssDNA-reporter was used (length: 32 b, 5’ FITC and 3’ Biotin). 20 µL of diluted reporter was mixed in 80 µL Assay buffer in a 96-well plate. HybriDetect was placed contemporaneous into wells using the 12-Strip-Dip-Template. Results are read after 3 minutes.
We found that a range of 0.2 – 2.0 pmol per LFA is good for an almost complete T-line elimination. Comparable C- and T-line intensities are generated for 0.02 pmol and 100 pmol reporter per LFA, although the best result was achieved in between these two concentrations. This apparently surprising result can be explained by a typical immunoassay associated phenomenon which also occurs in lateral flow tests: the high dose hook effect.
CAUTION: too much reporter causes the High Dose Hook Effect
The high dose effect is a typical immunoassay related phenomenon. It occurs, if too many LFA-relevant labels (Biotin / FITC / FAM / DIG) are introduced into the LFD. The general mechanism is based on a limited number of label-specific binding sites in the lateral flow system. If the number of relevant labels exceeds the number of available binding sites in the lateral flow system, the overall number of signal-creating “molecular sandwiches” will be reduced. A decreasing C-line and increasing T-line intensity is the result of a significant reporter excess. This is an important and characteristic limitation for immunoassay formats like LFAs or ELISAs. The following figure illustrates the reporter-induced hook effect in the HybriDetect system.
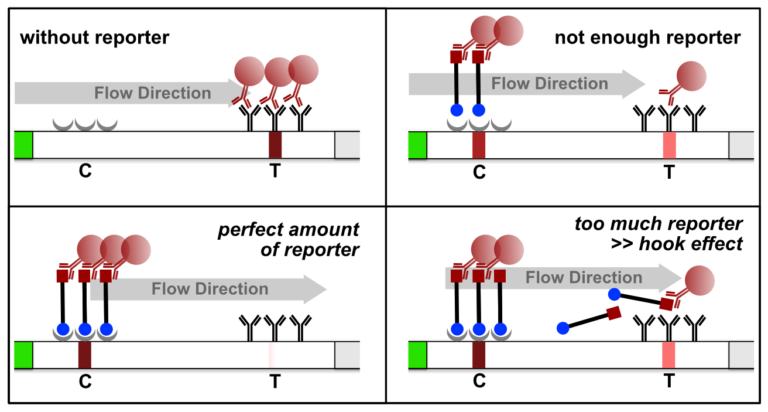
Figure 6. Reporter induced Hook Effect in the HybriDetect system.
It is necessary to understand the hook effect, to avoid misinterpretation and handling errors. According to this effect it is possible to create identical signal intensities with different concentrations of the dual labeled reporter. Furthermore, it is possible to create false negative results especially for assays, which are designed according to the described label incorporation strategy. It is crucial to limit the number of relevant labels in the LFA.
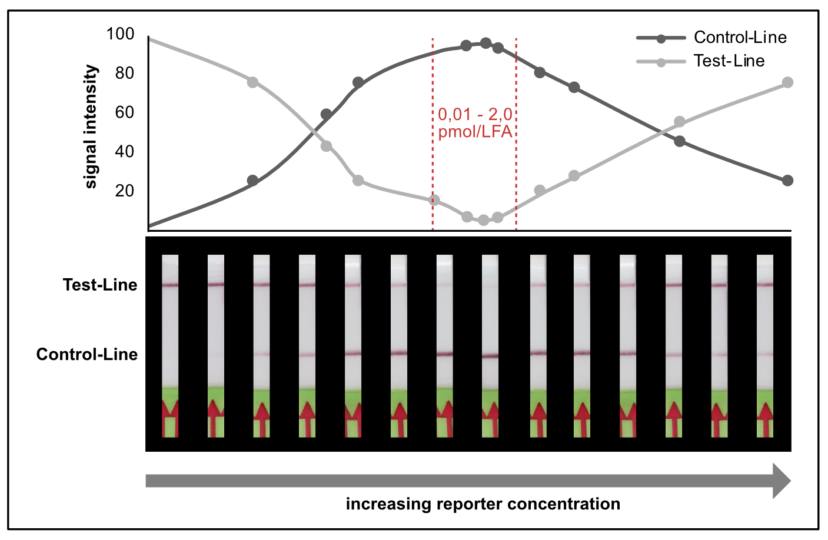
Figure 7. Idealized C- and T-line intensity depending on the reporter concentration in the LFA to illustrate the reporter induced hook effect in the HybriDetect system.
In other experimental settings we tried to determine the maximum number of labels per LFA without running into the hook effect. The following table gives a brief overview of the discussed limitations.
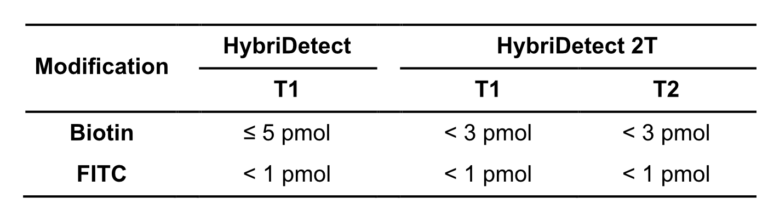
Table 2. Overview of the maximum label amounts before running into the hook effect. T-Lines are defined according to the instruction for use, and not according to CRISPR/Cas-related HybriDetect usage.
Tips and tricks for sufficient T-Line Elimination
# Good range for reporter concentration: 0,1 – 2 pmol / LFA
# Too little amount of reporter and too much reporter will result in increasing T-Line intensity >> difficult to interpret
# CAUTION >> Too much reporter induces high dose hook effect
# Long reporter sequence can result in secondary structures, or reporter-reporter interaction >> possible influence on the detectability
# Pretest new reagents and new lots
Advanced Tips
The use of 1 or 2 pmol may not be the optimum for a quick and efficient measurement of nuclease activity. If the reporter concentration in the nuclease assay deviates from T-line elimination range, the T-line intensity will be too strong for an intuitive interpretation. But there are some valuable tricks to achieve the best possible readout.
SPIKE IN: additional non cleaved reporter can be added to the Assay Buffer. Generally, you can use ssDNA reporter and increase stability of spiked assay buffer independent of the nuclease assay type. This is an easy tool for test-line intensity finetuning, if the reporter amount is not sufficient for T-line elimination. Assay buffer modification is an underrated tool in HybriDetect associated analysis.
DILUTE / less SAMPLE: at the end it’s all about pretesting your test system in the LFA context. Try to be confident using less sample or dilute before LFA. Less can be more!
SAMPLE on CRP: Try to applicate your sample directly on the Conjugate Release Pad (max. 10 µL) before you dip the stick into the assay buffer (in addition to the reporter SPIKE IN). The potentially cleaved reporter will have an advantage to bind to the mobile antibodies. Sometimes this can be beneficial to achieve sensitive test results.
TIME: Analysis time is extremely important. Two – three minutes of Lateral Flow are most frequently used in the reporter degradation dependent methods until documentation. The contemporaneous analysis start is important and can be challenging with 10 or more strips. To achieve comparable results regards analysis time, try to implement the 12-Strip-Dip-Template (can be downloaded here) in your assay procedure.
References:
- Myhrvold C et al., 2018. Field-deployable viral diagnostics using CRISPR-Cas13.
- Abudayyeh OO et al., 2019. Nucleic Detection of Plant Genes Using CRISPR-Cas13.
- Sullivan et al., 2019. Rapid, CRISPR-based, Field-Deployable Detection of White Spot Syndrome Virus in Shrimp.
- Zhang et al., 2020. A protocol for detection of COVID-19 using CRISPR diagnostics.
- Metsky et al., 2020. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design.
- Kaminsky et al., 2020. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection.
- Broughton et al., 2020. CRISPR–Cas12-based detection of SARS-CoV-2.
- Tsou et al., 2019. A CRISPR Test for Detection of Circulating Nuclei Acids.
- Zhang Y. et al., 2020. Evaluation of CRISPR/Cas12a-based DNA detection for fast pathogen diagnosis and GMO test in rice.
- Zhang Y and Ying YY, 2015. Homogeneous Immunochemical Assay on the Lateral Flow Strip for Measurement of DNase I Activity.
Author
André Breitbach
abreitbach@milenia-biotec.de
+49 (641) 948883 – 0