03/27
2020
6 new Articles, referring to the use of CRISPR/Cas and HybriDetect
DNA and RNA Detection with the Help of HybriDetect
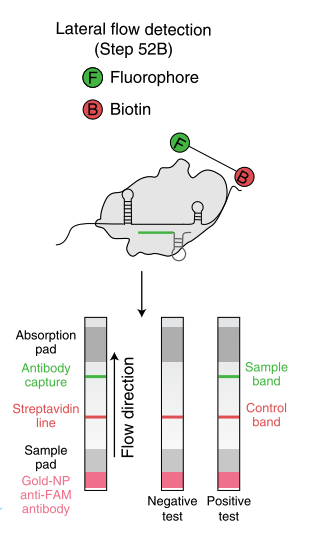
New techniques have been developed for the specific detection of DNA or RNA sequences. They all have in common, that they use CRISPR/Cas12 or Cas13. The enzyme recognizes a specific target sequence. After binding to this sequence, its endonuclease activity is activated and it cleaves any reporter sequence. This technique can be easily combined with our Lateral Flow Assay HybriDetect for the visualization of the results.
1. SHERLOCK: nucleic acid detection with CRISPR nucleases
read full article
Researchers from the Broad Institute recently published an article about a CRISPR-based diagnostic platform that combines nucleic acid pre-amplification with CRISPR-Cas enzymology for specific recognition of desired DNA or RNA sequences. The described technique is named the SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) method. This method can easily be combined with our HybriDetect for fast visualization of the results.
The CRISPR/Cas-enzymes Cas12 or Cas13 are activated by binding to a specific target. When activated, the endonucleases are able to cleave nonspecific reporters.
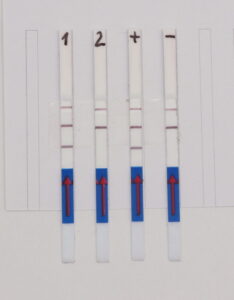
A simple, instrument-free and portable way to readout the SHERLOCK reaction is using our LFA (lateral flow assay) HybriDetect.
The Way HybriDetect is used in the SHERLOCK Assay:
A reporter labeled with FITC on one side and Biotin on the other side has to be used. Upon activation of Cas12/Cas13 this reporter is cleaved. The biotin side of the reporter is captured by the lower band of the HybriDetect dipstick. Anti-FITC antibodies, which are labeled with gold nanoparticles, will bind on the other side of the Reporter (FITC). A dark purple line is formed at the Control band of the dipstick. Upon activation of Cas12/cas13 by binding the specific target sequence the reporter is cleaved. The gold NP-labeled antibodies, which are bound to the FITC side of the reporter will travel across the test band of the dipstick. Here they are bound by species specific antibodies and form a purple line at the 2nd position.
Reference:
Kellner M. J., Koob J. G., Gootenberg J. S., Abudayyeh O. O., Zhang F., SHERLOCK: nucleic acid detection with CRISPR nucleases. Nature Protocols (2019); V 14 (10, 2986-3012)
2. SARS-CoV-2 Detection
read full article
Due to the current COVID-19 outbreak, many labs are working on fast and easy, but still accurate ways to detect SARS-CoV-2. Since now, the EUA-approved assay developed by the US CDC is the standard test for SARS-CoV-2, it is a qRT-PCR and needs 4-6 hours.
Broughton et al. developed a CRISPR-Cas12 related assay for the detection of SARS-CoV-2 from patient RNA in 30 minutes, called DETECTR.
Upstream a reverse transcription (RT) with a Loop-mediated Isothermal Amplification (LAMP) simultaneously (RT-LAMP) has to be done. After this, Cas12 detection of specific viral sequence and cleavage of a reporter molecule is carried out. The final step is the visualization by our lateral flow test HybriDetect. Which has the main advantage of showing quick results without any instruments needed.
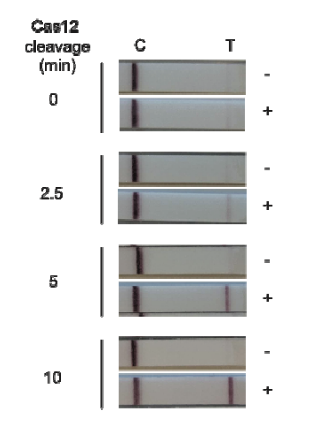
Clinical Testing of the DETECTR CRISPR Assay is currently ongoing.
Reference:
Broughton J. P., Deng X., Fasching C.L., Singh J., Streithorst J., Granados, A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C., Guevara H., Wadford D., Chen J., Chio C.Y., Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. medRxiv (2020); V 415
3. Detection of red tide dinoflagellate
read full article
Karenia mikimatoi is a common red tide alga, which can induce the death of fish and shellfish and therefore causes economic losses. The authors developed a new detection method using a LAMP combined with our HybriDetect for visualization. The whole procedure takes only 23 minutes from preparation of the sample to result.
Only a simple water bath or heat block is needed. The LAMP combined with HybriDetect shows is more specific and faster, compared to agarose gel electrophoresis or simple adding of SYBR green. The authors could detect the results 3 minutes after inserting the dipstick into their probe.
Reference:
Huang H. L., Gao W.F., Zhu P., Zhou C.X., Qiao L.L., Dang C.Y., Pang J.H., Yan X.J., Molecular method for rapid detection of the red tide dinoflagellate Karenia mikimotoi in the coastal region of Xiangshan Bay, China. Journal of Microbiological Methods (2020); V 168
4. Cas9-Mediated Lateral Flow Nucleic Acid Assay
read full article
Wang et al. presented a new method called CRISPR/Cas9-mediated lateral flow nucleic acid assay (CASLFA), which is one of the most convenient analytical techniques for analyzing immune response. They identified Listeria monocytogenes, GMOs and African swine fever virus (ASFV) with a detection limit of hundreds of copies of genome samples with high specificity within one hour on our HybriDetect. ASFV infected samples could be detected with an accuracy of 100% compared to RT-PCR.
The authors point out, that no laboratory environment is needed. The point-of-care-use without any technical expertise and ancillary equipment is given with this new technique.
Reference:
Wang X., Xion E., Tian T., Cheng M., Lin W., Wang H. Clustered Regularly Interspaced Short Palindromic Repeats / Cas9- Mediated Lateral Flow Nucleic Acid Assay. ACS Nano (2020), IF 13.903.
5. Detecting Plant Genes
read full article
Abudayyeh et al. presented a modified SHERLOCK detection method to quantify levels of the glyphosate resistance gene in a mixture of soybeans and to detect multiple plant genes in a single reaction. They described a field-ready SHERLOCK platform combined with Milenias HybriDetect for detection of genes in a range of agricultural applications.
Prior the Cas-reaction (20 minutes), a Recombinase Polymerase Amplification (RPA), which takes 10 minutes, is done.
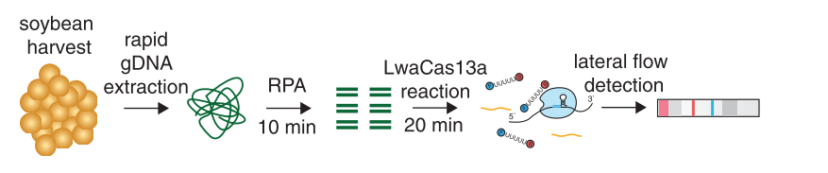
The high sensitivity (single molecule, 2 aM input concentration in 1 µL sample) and specificity for single nucleotide discrimination is the major advantage of this technique. It can be used for field application, and easily combined with HybriDetect when a reporter labeled with biotin and FAM is added to the SHERLOCK reaction.
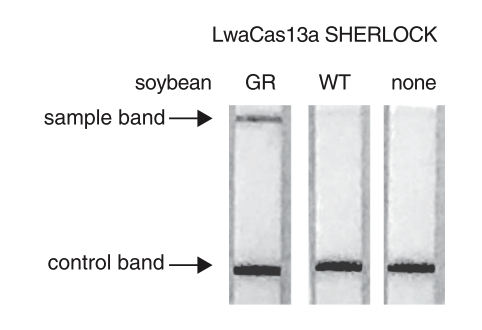
Taken together the described method is a useful platform for many biotechnological and agricultural applications.
Reference:
Abudayyeh O. O., Gootenberg J. S., Kellner M. J., Zhang F., Nucleic Acid Detection of Plant Genes Using CRISPR-Cas13. The CRISPR Journal (2019), V 2 (3, 165-171)
6. Detecting Pathogens and GMOs
read full article
Limit of rapid, low cost, user-friendly, field deployable DNA test method
Another plant application using CRISPR and HybriDetect is described by Zhang et al.. The group used a Cas12 based DNA detection method for crop disease diagnosis and a GMO test. They also used a RPA combined with Cas12 cleavage and HybriDetect for visualization.
A great advantage of this method is that it runs at 37°C and no extra instruments except filter paper and HybriDetect is needed.
The tested rice blast pathogen and Bt-rice (transgenic rice) were efficiently identified from leaf disc samples.
The described method is a rapid, low-cost, user-friendly DNA test method, which can be easily applied in field for crop disease diagnosis and GMO administration.
Reference:
Zhang Y., Zhang Y., Xie K., Evaluation of CRISPR/Cas12a-based DNA detection for fast pathogen diagnosis and GMO test in rice. Molecular Breeding (2020); V 40,11
Click here to see how to perform HybriDetect
Author
Dr. Jacqueline Hoffmann
QA — RA
jhoffmann@milenia-biotec.de
+49 (641) 948883 – 0

