Intro Solisbiodyne

PCR, qPCR and RT-qPCR reagents
Solis BioDyne has been developing and producing life science reagents since 1995, having become one of the leading reagent suppliers in Europe today. High standards for production and service have made Solis BioDyne a trusted trademark worldwide. Our DNA polymerases, PCR Master Mixes, qPCR Mixes and reverse transcription reagents are used by a quickly growing number of customers across the globe, including top research institutes and biotech-companies. Solis BioDyne has partners in both private and state sectors, with cooperation projects ranging from OEM production to scientific research.
Product Categories
Endpoint PCR
qPCR and RT-qPCR
cDNA Synthesis
Additional Enzymes and Reagents
Bisulfite Conversion for Illumina Methylation Arrays
Bisulfite Conversion for Illumina Methylation Arrays
Troubleshooting & Best Practice Guidelines
Methylation arrays are common platforms for analyzing 5-methylcytosine. The Infinium™ MethylationEPIC BeadChip and Infinium™ HumanMethylation450 BeadChip (commonly referred to as the EPIC and 450K arrays respectively) as well as the recently launched Infinium™ Mouse Methylation BeadChip from Illumina® all utilize Zymo Research’s bisulfite conversion technologies to distinguish 5mC from unmodified cytosines. In addition to the test probe sets, the arrays include bisulfite conversion quality control probes. In some instances, the analysis software will flag samples for low bisulfite conversion efficiency. Possible causes for these warnings are low bisulfite conversion efficiency, low DNA input/quality, and chip failure.
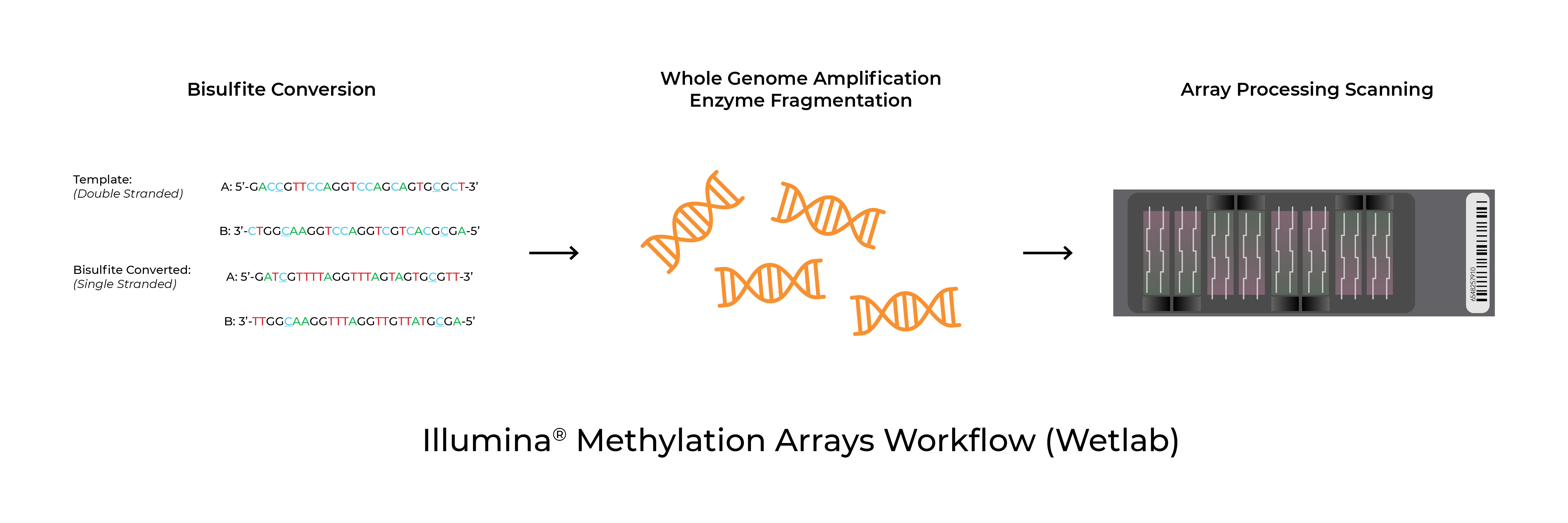
Validated Protocols
The only approved and validated bisulfite conversion kit for the Infinium™ Methylation arrays is the EZ DNA Methylation kit (catalog numbers: D5001, D5002, D5004). It is critical to follow the Illumina® recommended incubation protocol (16 cycles of 95°C for 30 seconds, 50°C for 60 minutes). Additional details can be found in the appendix of the respective bisulfite conversion protocols. While other bisulfite conversion kits will result in high-quality bisulfite converted DNA, only samples processed using the validated kits are supported by Illumina®.
Along with using the validated bisulfite conversion kits, it is important to follow the rest of the BeadChip protocol as written, including the downstream software analysis. Illumina® periodically reviews the performance of the analysis software and probe performance. Use the most up-to-date software and manifest files to ensure that downstream analysis is correct.
DNA Input & Quality

The amount and quality of genomic DNA used in these assays is critical to success. For best results, DNA should be as intact and high quality as possible. The arrays rely on a signal to noise ratio to determine the conversion efficiency. When low input or low quality DNA is used, the signal to noise ratio is much lower and can affect the reliability of the bisulfite conversion control signals.
The required minimum amount of DNA is 250 ng for the manual protocol and 1000 ng for the automated protocol. Genomic DNA should be quantified using a dsDNA specific method such as Picogreen® or Qubit®. NanoDrop® or other spectrophotometric methods are not recommended for quantification due to issues with successfully distinguishing DNA from RNA. RNase treatment can also help ensure that DNA quantification is accurate.
When working with DNA isolated from FFPE samples or other degraded DNA samples, 500 ng DNA inputs or higher are highly recommended. Additionally, single-column bisulfite conversion (vs. the 96-well plate) is recommended for degraded samples due to the ability to use smaller elution volumes. Bisulfite-converted FFPE DNA should then be treated with the Illumina Infinium™ FFPE DNA Restoration Kit (as described in the manual) before processing the array. The entire sample should be used.
Low Bisulfite Conversion Efficiency

Bisulfite conversion can be impacted by a variety of factors. The quality of the CT Conversion Reagent is critical for successful conversion. It is recommended to prepare the reagent fresh before each conversion if possible. Otherwise, the prepared reagent should be stored according to the guidelines listed in the protocol. The conversion reagent should not be exposed to light or oxygen any more than necessary. If processing 96-well plates, a multichannel pipette should be used and the prepared conversion reagent should be added last to the plate.
Conversion should be performed in a thermal cycler with a heated lid. Samples and conversion reagent should be mixed thoroughly (no observed mixing lines) and fully spun down before placing tubes in the thermal cycler. If the tubes are not fully spun down or the lid is not heated properly, precipitation may form around the lid of the PCR tube. After the incubation, any precipitation that is observed should be avoided when transferring the sample as it could contain unconverted DNA.
The desulphonation incubation should be stopped at 15 minutes (20 minutes is the absolute maximum). Leaving the desulphonation buffer on the column longer than the recommended time can result in additional degradation to the sample.
A bisulfite conversion quality control check is highly recommended. Bisulfite conversion can be assessed using a variety of methods. Bisulfite converted DNA can be quantified by qPCR or using the RNA setting on NanoDrop® (or similar spectrophotometric method). The expected DNA yield after bisulfite conversion is approximately 70-80%. This quantification step will help ensure that there is enough bisulfite converted DNA recovered for downstream processing. Bisulfite conversion efficiency can be assessed via colony Sanger Sequencing or via probe/TaqMan® assay. In rare instances, sample purity may be responsible for low bisulfite conversion efficiency. In these cases, re-extraction or further cleanup is recommended.
Chip Failure
On occasion, chip failures may occur. If multiple samples on a single chip have low bisulfite conversion efficiency as determined by the BeadArray Controls Reporter, this might indicate a chip failure rather than a bisulfite conversion issue. In these instances, re-running leftover bisulfite-converted sample on a new chip can resolve the issue. To further troubleshoot, a post-bisulfite conversion/ pre-array QC is helpful to determine whether to rerun the samples.
Chip Failure
Methylation arrays such as the 450K and EPIC array are helpful tools for analyzing methylation changes in a variety of human and mouse samples. Whenever a sample is flagged for low bisulfite conversion, it is best to confirm that the following is true for your project:
- ●Validated and updated protocols were used
- ● DNA was as high-quality and intact as possible. Higher DNA input amounts were used for fragmented/low-quality samples
- ● Bisulfite conversion was performed according to best practices
Following these simple steps will help determine the reason for the bisulfite conversion probe failure.
| DNA Methylation Analysis | |
| Bisulfite Methods | |
| Bisulfite conversion | EZ DNA Methylation Kits |
| NGS platforms | Pico Methyl-Seq Library Prep Kit |
| PCR platforms | |
| Bisulfite Free Methods | |
| Antibody Methods | |
| Enzymatic Methods | OneStep qMethyl Kits |
| Chromatin Analysis | |
| ChIP | |
| Chromatin Structure | Zymo-seq ATAC |
February 11, 2021
Epigenetic clock, Zymo leading the way
Epigenetic clock, Zymo leading the way
With the increasing number of refugees seeking to settle in the European Union, and with potential benefits for refugees being significantly increased for minors, European authorities are calling for better methods of verifying an individual’s age when reliable records are unavailable. Zymo Research, through an exclusive license to the Horvath Epigenetic Clock, is leading the way to a fast, reliable, commercially available epigenetic age test.
Can epigenetics help verify the age claims of refugees?
When local authorities in Hildesheim, Germany, didn’t believe an asylum seeker who claimed to be under 18 years old — and thus eligible for privileged treatment — police turned to a blood-based age test sold by a California company.
In a paper published online in May, a team led by forensic-medicine specialist Stefanie Ritz-Timme of the University of Dusseldorf in Germany said that these tests were not ready for use in sensitive forensic evaluations.
But now, in the charged political atmosphere that has accompanied the arrival of millions of refugees to Europe, forensic scientists across the continent are joining forces to improve epigenetic-clock-based tests — with a focus on whether they might be used to help determine the age of refugees whose claims to be under 18 are disputed. They hope that, with time, such tests could replace existing methods, which assess the maturity of bones or teeth to determine an individual’s age but are imprecise and can be controversial.
Photo Credit: Nemanja Pancic/SIPA/Shutterstock
Is an Epigenetic Clock the Key to Asylum?
Is an Epigenetic Clock the Key to Asylum?
A transcript of BBC World Service Radio’s Newsday interview with Keith Booher, PhD, of Zymo Research Corporation about their DNAge® test being used to determine the age of refugees seeking asylum in Europe as minors.
BBC: Could a finger prick test for blood make it easier for child refugees to have their age verified? That’s a question that California scientists have been thinking about. The field is known as Epigenetics and the test involves examining chemically modified DNA to create a sense of how old someone actually is. Keith Booher is a scientist in California and he has been part of the development of this device.
Booher: What we’re simply trying to do is to make an age determination or quantify aging. For example, in forensics cases when the provenance or age of a sample is unknown, we can make that assessment. There’s also some indications that it may be useful for biologically aging someone – so they may have been born 45 years ago, and are 45 years old but based on certain life decision they make – the diet they choose to eat, if they get enough exercise, do they live a stress-free lifestyle? Their biological age may be older or younger than their chronological age, just depending on the lifestyle they lead. And we should be able to quantify that as well.
BBC: So if I asked you now, how old you are, Keith, what would you tell me?
Booher: I’m 38 years old.
BBC: But that’s the chronological age, is that right?
Booher: Correct!
BBC: Which one is more important, biological or chronological?
Booher: You know, if you’re a young teenager trying to get your driver’s license, obviously your chronological age is very important. But if you are in advanced years of your life or you may have made some choices that are… poor when you were younger or over the course of your life, then biological age becomes much more critical.

BBC: How did you end up using your science with refugees?
Booher: Our information was passed to law enforcement officials working in Germany. And they had a case where they were trying to accurately determine the age of a refugee asylum seeker and they determined that the methods available to them at that time were not precise enough to get a good age determination and they reached out to us and asked if we could help.
BBC: What have you found in the course of your work? How accurate is your science at the moment?
Booher: Currently, epigenetic clocks are the most accurate, most precise way to age a sample. So our platform that we offer at Zymo Research has a median error of under two years – it’s about one and a half years. Other publications, people using different types of methods – you know, similar epigenetic aging, have shown median errors anywhere between two to four years. On aggregate, you have a precise age estimation method.
BBC: Has it helped clarify or clear doubt about age?
Booher: Yeah, I think so. In the case of the asylum seeker, I was told this person presented with no official documentation. The person claimed to be a minor, and therefore entitled to certain protections and assistance according to United Nations guidelines. So in this case, I think it definitely helped clear up any doubt regarding the age of the subject.
BBC: I can see it working with football and other sports as well. You know, where there have been all sorts of questions about ages.
Booher: Yeah, that’s correct! That’s a good point. So there’s certain rules about the age of participants engaged in these activities in different leagues.
BBC: Keith Booher from Zymo Research Corporation which is developing Epigenetics.
References:
Alexandra, K. (Producer). (2018, September 24). Newsday [Radio Broadcast]. London, UK: BBC World Service Radio
A Piece of the Puzzle
A Piece of the Puzzle
Autism (autism spectrum disorder) is one of the most common lifelong neurodevelopmental disorders, affecting an estimated 1 in 59 children in the United States 1. Due to its prevalence, autism has become a heavily researched topic in the field of genomics, as medical professionals and advocacy organizations look for a deeper understanding of its wide spectrum of symptoms. Genome-wide studies have suggested the involvement of hundreds of genes and genetic pathways in the development of the disorder. An increased risk of autism development is also known to be influenced by environmental factors such as premature birth, oxygen deprivation, infection during pregnancy, and parental age at time of conception 3. Recent studies have shown that these factors influence what is known as our epigenetics.
But What is Epigenetics?
Epigenetic regulation – in the form of DNA methylation, histone modification, and chromatin remodelling – helps govern the proper expression of genes in the genome. One’s environment continuously shapes their epigenetics. Factors that influence epigenetics can include one’s diet, exercise, stress level, drug abuse, and exposure to toxic pollutants. Failure in epigenetic regulation can result in aberrant gene expression (mutations, deletions, copy number aneuploidies) that contributes to halted neural development 4. Thanks to progresses in Next-Gen Sequencing, scientists have been able to map over 600 human epigenes associated with neurodevelopmental disorders, including autism.

Ongoing Epigenetic Research
Autism research is a continuously growing field, especially in the context of epigenetics. Currently, diagnosis and subsequent treatment is tough due to the degree of heterogeneity of traits those with autism can have. Fortunately, as sequencing technology becomes cheaper, scientists have more opportunities to analyze the epigenome with higher resolution. Without a high price point necessary for entry into autism research, smaller labs now have an opportunity to make significant discoveries.
There are many ongoing studies that aim to understand the epigenetic mechanisms behind the pathogenesis of autism spectrum disorder, as well as possible methods for treatment. A recent exciting study published in Nature Neuroscience demonstrated that mouse models of autism could be rescued via inhibition of histone deacetylase 8. These mice were deficient in the SHANK3 gene, another high-risk target linked to autism spectrum disorder. Mutations in the gene results in dysregulation of emotional and cognitive response in both humans and mice. The researchers found that low-dose treatment by anti-cancer drug romidepsin, a histone deacetylase inhibitor, fully restored gene function and completely reversed the social deficits of the mice after 3 days. This effect lasted 3 weeks in the mice, equivalent to the length of many years in humans.
Why Autism Research Is Important
Research that continues to find links between autism and epigenetics provides clues to the cause and mechanisms of the disorder, as well as invaluable biomarker discoveries. While it is currently thought that there is no single underlying cause of autism, epigenetic biomarkers can be used for risk assessment prior to diagnosis, diagnosis of the disorder itself, or even provide insight into its severity 9.
Studies show that early intervention leads to positive outcomes later in life for people with autism 1. By furthering research on such a prevalent disorder, we can better prepare families at an earlier timepoint. Considering autism’s broad range of characteristics, scientific findings into the disability may also provide insight into the inner workings of other related neurodevelopmental disorders.
Citations:
1. https://www.autismspeaks.org/what-autism
2. Hans van Bokhover. “Genetic and epigentic networks in intellectual disabilities”. Annual Review of Genetics, 2011. 45: 81-104
3. https://www.autismspeaks.org/environmental-factors-autism
4. Michelle Siu and Rosanna Weksberg. “Epigenetics of Autism Spectrum Disorder”. Neuroepigenomics in Aging and Disease, 2017. 978: 63-90
5. Luikenhuis, S., et al. “Expression of mecp2 in postmitotic neurones rescues rett syndrome in mice”. Proc Natl Acad Sci USA, 2004. 101: 6033-6038
6. Samaco, RC., et al. “Epigenetic overlap in autism-spectrum neurodevelopmental disorders: mecp2 deficiency cuases reduced expression of UBE3A and GABRB3”. Hum Mol Genet, 2005. 14: 483-492
7. Wong, C., et al. “Epigenome-wide DNA methylation analysis of monozygotic twins discordant for diurnal preference”. Twin Res Hum Genet, 2015. 18: 662-669
8. Qin, L., et al. “Social deficits in shank3-deficient mouse models of autism are rescued by histone deacetylase inhibition”. Nature Neuroscience, 2018. 21: 564-575
9. Loke, Yuk Jing, et al. “The role of epigenetic change in autism spectrum disorders”. Front Neurol, 2015. 6: 107
Where There’s a Whale There’s a Way
Where There’s a Whale There’s a Way
Their songs haunt the deepest depths of the ocean and their behaviors have intrigued scientists for generations. The humpback whale itself is a mystery in many ways, such as having no real visible age indicators after they are one year old.
Studying the age characteristics of sensitive animal populations can provide valuable ecological insights into questions of reproductive success, lifespan, survival trends, and more. In recent decades, marine biologists have used different methods to measure age in certain whale species. However, most have notable limitations.
Limitations
For example, the age of deceased whales can be estimated using various invasive biopsy techniques, but doing so precludes the monitoring of living subjects over time. Alternatively, researchers attempted to measure the age of live whales using telomeric genetic markers, but those methods suffer from a lack of specificity and too much technical noise. By contrast, a number of recent research articles demonstrated that DNA methylation-based age estimators, commonly referred to as epigenetic clocks, are a highly accurate and technically robust means to measure aging in mammalian species as diverse as mice, canines, humans, and other primates1-3.
A group of researchers in Australia now show that DNA methylation analysis is an effective way to help predict the age of living whale populations, thus providing a new tool to study the demographics of these majestic ocean leviathans.
In their study, Polanowski et al.4 noted the development of DNA methylation-based age clocks and set out to develop a similar test for use in humpback whale (Megaptera novaeangliae) research. They focused on the evolutionarily conserved 5’ regulatory regions of genes whose changing DNA methylation patterns correlated with age. The authors first generated a calibration data set using a cohort of 45 humpback whales originating primarily from the Gulf of Maine and with known ages ranging from a few months up to 30 years. Importantly, the tissue sample source used in the analysis came from minimally invasive skin punch dart biopsies.

Humpback Epigenetic Age Assay (HEAA)
The scientists called their new experimental tool the Humpback Epigenetic Age Assay (HEAA). Application of their 3-gene test demonstrated a high positive correlation (R2 = 0.787) between DNA methylation change and age. Furthermore, a leave-one-out cross validation from the calibration cohort data set demonstrated precision and accuracy (Mean difference = 3.75 years; Standard deviation = 2.991 years) between the predicted and known ages of the samples.
Having established the HEAA, Polanowski et al. then set out to test their model on a humpback whale population of unknown age composition. Using skin biopsies once again, they measured the epigenetic ages of 63 humpback whales migrating north near Evans Head, Australia, and observed an average population age of 10 years, with a total age range from 0 to 52 years.
The authors further noted that the distribution of the measured ages resembled what would be expected for a sample population of that size and in that region after comparing their dataset to one obtained in the same area 50 years earlier, before the effects of whaling caused the fishery to collapse in the 1960s. One final test of the HEAA showed its ability to correctly pick the ordinal age-relationship between parent and progeny in both data simulations (<90% accuracy) as well as an empirical test of mother-calf pairs taken from the test and calibration data sets, respectively. The assay proved accurate in all 12 pairings tested.
With the HEAA, Polanowski et al. present an epigenetic clock similar to what has been shown to work so effectively in other mammalian species. Initial calibration and testing of the HEAA demonstrated its broad precision, accuracy, robust nature, and compatibility with a minimally invasive and readily obtainable sample source. Continued use of the HEAA, or similar DNA methylation-based age estimators, will help scientists track the overall health of whale populations well into the 21st century.
Citations:
[1] Horvath S. DNA methylation age of human tissues and cell types. Genome Biology. 2013;14(10): R115. doi:10.1186/gb-2013-14-10-r115.
[2] Petkovich DA, Podolskiy DI, Lobanov AV, Lee SG, Miller RA, and Gladyshev VN. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metabolism. 2017;25(4);954-960.e6. doi:10.1016/j.cmet.2017.03.016.
[3] Thompson MJ, vonHoldt B, Horvath S, and Pellegrini M. An epigenetic aging clock for dogs and wolves. Aging. 2017;9(3):1055-1068. doi: 10.18632/aging.101211.
[4] Polanowski AM, Robbins J, Chandler D, and Jarman SN. Epigenetic estimation of age in humpback whales. Mol Ecol Resour. 2014;14(5):976-87. doi: 10.1111/1755-0998.12247.
Long-Read Epigenetics with Microbiome Standards
Long-Read Epigenetics with Microbiome Standards
Is m6A the new 5-mC?
Of the several chemical and structural modifications to DNA that are known to influence gene expression, 5-methylcytosine (5-mC), has garnered the most attention because of its role in transcriptional silencing and the readily available techniques to investigate it. For instance, bisulfite sequencing via short-read Next Generation Sequencing (NGS) has become the gold-standard for 5-mC detection due to its single-nucleotide resolution and the ever-decreasing cost of whole genome sequencing. The pairing of bisulfite to NGS has greatly expanded our understanding on the extent of cytosine methylation influence on gene expression.
But, the application of NGS technologies to analyze the genomes of complex microbial communities (e.g. microbiome and metagenomic samples) has caused other DNA modifications to gain more attention. For example, N6-methyladenine (m6A) plays important roles in bacterial survival and interactions with hosts 1. Like other epigenetic base modifications, m6A contributes to the regulation of gene expression as well other house-keeping functions in bacteria. Unfortunately, methylation detection by short-read bisulfite sequencing is based on cytosine to uracil conversion and cannot detect adenine modifications, such as m6A.
This is why many researchers are turning to long-read sequencing platforms (3rd generation sequencing technology), since they use differences in electronic signals as bases to pass through nanopores and detect other types of base modifications. Like any new technology, methylation detection with 3rd generation sequencing needed benchmarking with the use of well-defined standards and controls.
Not one, not two, but four sequencing platforms!
While the use of one sequencing platform is sufficient for most studies, some researchers incorporate two sequencing methods when verifying a new bioinformatics tool. However, to validate a new N6- methyladenine (m6A) detection tool named mCaller, McIntyre, et al. sequenced the ZymoBIOMICS Microbial Community Standard using PacBio, Oxford Nanopore, MeDIP-seq and whole-genome bisulfite sequencing 1. Single-molecule sequencing techniques, such as PacBio 2 and Oxford Nanopore Technologies (ONT) 3 have been used to detect m6A, but no method has had cross-validated results by using a well-characterized reference material on several sequencing platforms – until now.
Older m6A detection methods based on immunoprecipitation were limited in terms of nucleotide resolution, while previous single-molecule sequencing detection tools suffered from lower base modification calling (~70%) 3, 4. To improve accuracy, mCaller employs a neural network to learn and test different classifiers to detect m6A in Nanopore data generated from the E. coli MG1655 (a K-12 strain) genome.
To validate the tool, the bacterial genomes of the ZymoBIOMICS microbial community standard were sequenced via ONT, PacBio, and MeDIP-seq, and mCaller detection accuracy was compared across the different data sets. Remarkably, detection accuracy increased to 84.2% for high quality reads, and even 95.4% for single sites with at least 15x coverage, when compared to immunoprecipitation methods. Additionally, the methylome of the standard was verified with the use of the TruSeq DNA Methylation Kit.
Regulatory-Grade Genomes
As an additional control, the PacBio sequencing of the ZymoBIOMICS Microbial Community Standard was performed at two separate locations: the University of Florida and the Database for Reference Grade Microbial Sequences (FDA-ARGOS). The goal of FDA-ARGOS is to create a database of reference-grade microbial sequences available to the public. Because the sequencing of the ZymoBIOMICS Microbial Community Standards met the FDA-ARGOS quality requirements, they have been accepted as regulatory-grade genomes with the designations of FDARGOS_606 and FDAARGOS_612 under the BioProject number PRJNA477598.
“Overall, our results demonstrate a need for tool evaluation at a variety of sequence contexts, for which we propose the continued use of this well-validated microbial reference community.” – McIntyre, et al.
Team work makes the dream work
Although the ZymoBIOMICS Microbial Community Standard was developed as microbiome reference material, its well-defined and characterized composition has made it an excellent control for epigenetic sequencing. Such a cross-discipline use of reference materials and sequencing techniques demonstrates the power of inter-lab cooperation and the ability for different field leaders to push the capabilities of current technology.
References:
[1] McIntyre ABR, Alexander N, Grigorev K, Bezden D, Sichtig H, Chiu CY, Mason CE. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nature Communications. 2019 10 (579).
[2] Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Fengg Z, Losic B, Mahajan MC, Jabado OJ, Deikus G, Clark TA, Khai L, Murray IA, Davis BM, Keren-Pas A, Chess A, Roberts RJ, Korlach J, Turner SW, Kumar V, Waldor MK, Schadt EE. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nature Biotechnology 2012 30(12)
[3] Stoiber M, Quick J, Egan R, Lee JE, Celniker S, Neely RK, Loman N, Pennacchio LA, Brown J. De novo Identification of DNA Modifications by Genome-Guided Nanopore Signal Processing. BioRxiv 2017. doi: https://doi.org/10.1101/094672
[4] Rand AC, Jain M, Eizenga JM, Musselman-Brown A, Olsen HE, Akeson M, Paten B. Mapping DNA methylation with high-throughput nanopore sequencing. Nature Methods 2017 14(4)
April 15, 2020
CONTROLS FOR DNA METHYLATION ASSAYS
CONTROLS FOR DNA METHYLATION ASSAYS
How Controls Boost Reliability & Reproducibility
CONTROLS FOR METHYLATION ASSAYS: HOW, WHEN & WHY TO USE THEM
All scientific experiments need controls. Specific controls are required for certain type of experiments, such as DNA methylation assays. DNA methylation is a powerful, well-studied epigenetic mark that continues to provide extensive insight into transcription regulation, embryonic development, genome stability, and chromatin structure. Changes in DNA methylation profiles have also been linked to the development of many diseases 1. Many clinical applications have utilized DNA methylation as a biomarker for early detection, diagnostics, and personal therapy 2, 3. This requires accurate characterization of methylation patterns at a specific region or single-base level.
Most commonly used DNA methylation assays include bisulfite sequencing using NGS technologies, methylation-specific PCR (MSP), and methylation-sensitive restriction enzyme (MSRE) assays. To minimize bias in quantifying methylation levels, especially in clinical applications, each assay needs to be carefully designed and optimized to ensure its robustness4. Integrating established controls is key for optimizing assays and monitoring assay’s effectiveness . For optimization of DNA methylation assays, it is best to use established DNA standards with known methylation levels. Typically, this requires a methylated and non-methylated DNA standard, which contain nearly 100% and 0% methylation, respectively, at all CpG sites. The use of these two standards individually or in combination can help identify potential biases in assay development. For established workflows, these standards can serve as a positive and negative control to validate the procedures and results, which is essential in clinical testing to enhance quality control and assurance.
QUALITY CONTROLS FOR VALIDATING DNA METHYLATION ASSAY WORKFLOWS
As a good practice, methylated and non-methylated standards should be processed in parallel to experimental samples as a quality control for the whole workflow. In contrast to standards that are expected to produce a consistent result each time, experimental samples can have many variables, making troubleshooting difficult. In PCR-based and NGS-based DNA methylation assays, inhibitors carried over from the samples, primer design, enzymatic reagents, instrument failures, and many other factors can cause an assay to fail. Data produced by the standards can suggest where to start troubleshooting. For example, if standards perform as expected when run in parallel with experimental samples which perform poorly, the workflow is functioning properly, suggesting the experimental sample quality may be an issue. If standards did not perform as expected, there may be an issue within the workflow that caused the assay to fail.
CONTROLS FOR OPITIMIZING AND CALIBRATING BISULFITE-BASED ASSAYS
Bisulfite PCR (BSP) is a common methodology used for methylation analysis at single-base resolution. Samples are first bisulfite converted and amplified using bisulfite-specific primers. The PCR products generated are sequenced (i.e. Next-Gen Sequencing, Sanger Sequencing, pyrosequencing) and analyzed to determine the methylation status at every cytosine. However, it is critical that primer pairs designed for BSP amplify both methylated and non-methylated sequences with the same efficiency (Figure 1). Otherwise, any amplification bias can result in skewed methylation levels.
Using methylated and non-methylated DNA controls, BSP primers can be easily optimized (validated) based on following criteria:
- +Specific product amplification
- +Equally robust amplification of both methylated and non-methylated DNA standards
- +When sequenced, the methylated and non-methylated DNA standards should result in nearly 100% and 0% methylation, respectively. If the results deviate from the expected result, this may indicate PCR bias.
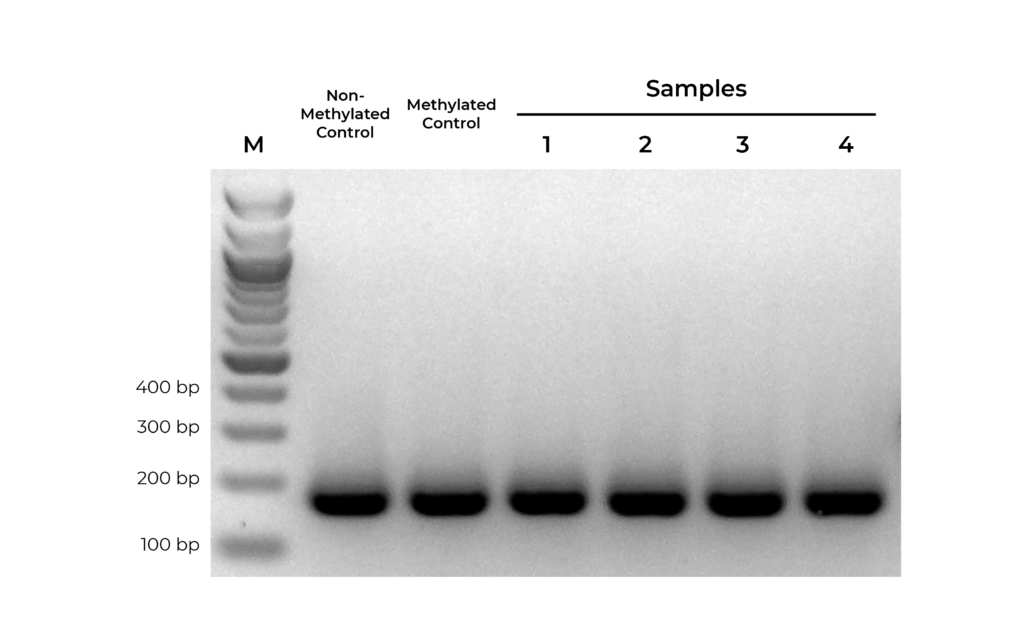
Figure 1. Bisulfite PCR primers should show equally efficient amplification of methylated, non-methylated, or partially methylated templates. Equal amounts of bisulfite converted non-methylated and methylated DNA controls were amplified in parallel with experimental samples using primers designed to target MGMT. Observed bands for methylated and non-methylated templates are of equal intensity, indicating that the primer pair is unbiased.
Unlike bisulfite PCR, methylation-specific PCR (MSP) requires methylated and non-methylated templates to be differentially amplified by two different primer sets. One is a methylation-specific primer set, and if amplification occurs, it indicates the amplified region is completely methylated. The second primer set targets non-methylated templates, so any amplification with this primer set indicates the amplified region is completely non-methylated. If amplicons are generated by both primer sets, the region is partially methylated.
Such two primer sets required for MSP can also be optimized and validated utilizing Methylated & Non-Methylated DNA Controls using following guidelines:
- #Specific product amplification
- #The methylated primer will only amplify the methylated control DNA and will not amplify the non-methylated DNA.
- #The non-methylated primer will only amplify the non-methylated control DNA and will not amplify the methylated DNA.
During primer validation, having characterized methylated and non-methylated DNA standards can streamline the process. For example, if a slight PCR signal is observed using the methylated standard with the non-methylated primers, or vice versa, it would indicate that the primer set is non-specific. For established workflows, especially in clinical assays, methylated and non-methylated DNA standards can serve as positive and negative controls to validate the MSP workflow. Standards will help confirm that the entire process – bisulfite conversion, amplification, and analysis – are functioning appropriately, and the results for the test samples are reliable (Figure 2).
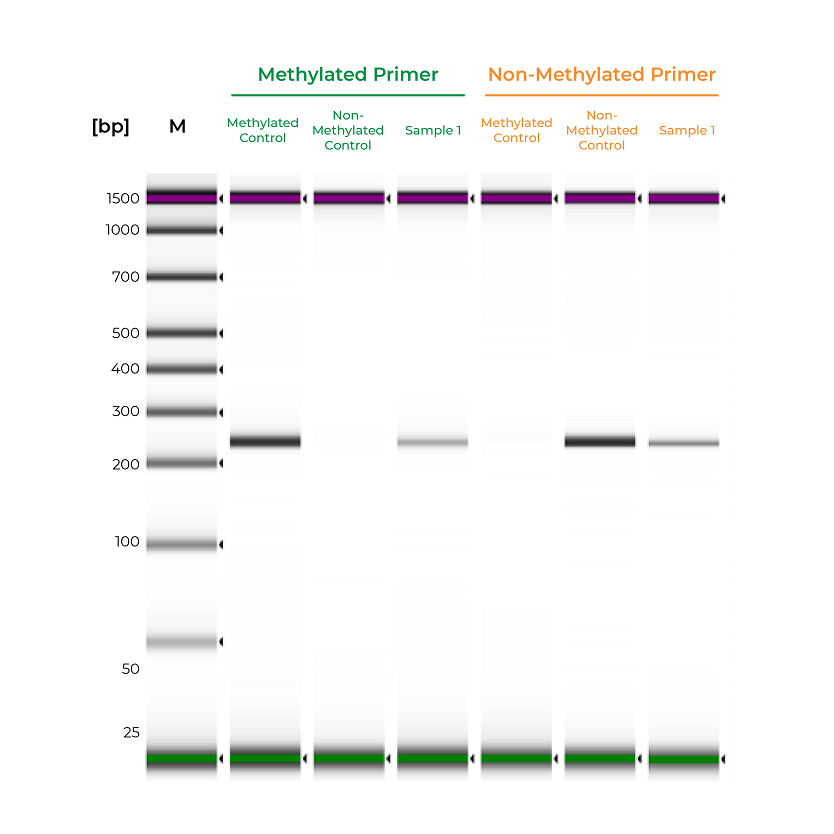
Figure 2. Methylated and non-methylated controls are important for validating the sensitivity of MSP assays. The methylated and non-methylated controls are only amplified by the methylated and non-methylated primers, respectively. Sample 1 is amplified by both primer sets, indicating mixed methylation. Samples were analyzed on the Agilent D100 ScreenTape.
In addition to validating primers, DNA standards can be used in place of precious samples to optimize the annealing temperature for new primer sets (Figure 3). An annealing temperature gradient should be performed for each newly designed primer set to ensure optimal amplification of the intended target.
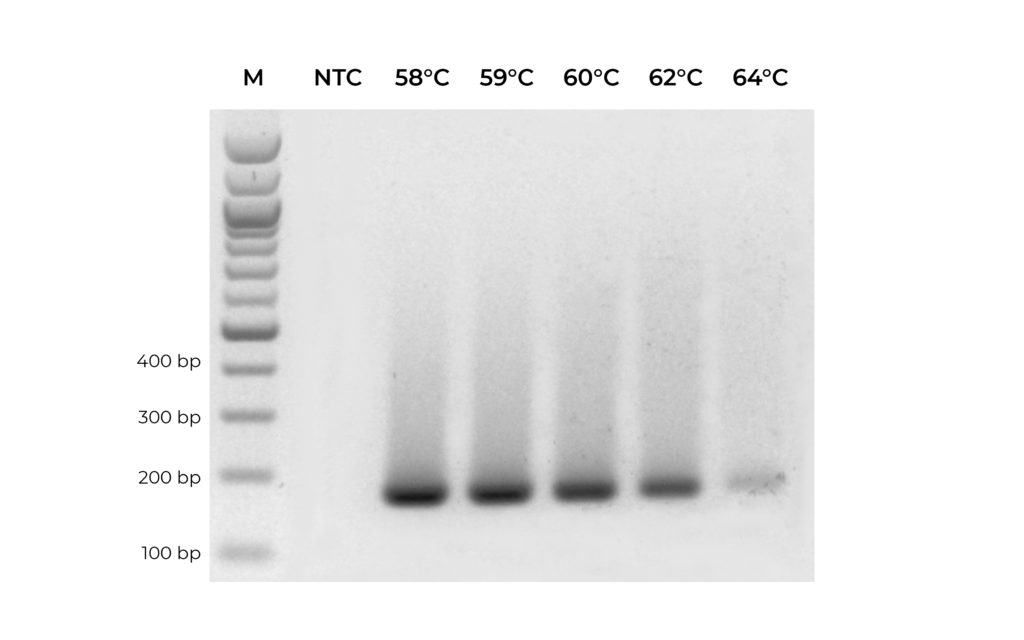
Figure 3. A primer set targeting MGMT was optimized by testing replicate PCR reactions with annealing temperature ranging from 58°C to 64°C.
CONTROLS FOR METHYLATION SENSITIVE RESTRICTION ENZYME (MSRE) ASSAYS
Methylation sensitive restriction enzyme (MSRE) assays rely on restriction enzymes being able to distinguish methylated versus non-methylated cytosines. If the restriction site is not methylated, the enzyme will digest the DNA while methylated sites will be uncut. Then, methylation at the site can be evaluated using quantitative PCR by comparing the amplification of the digested versus undigested sample. The smaller the ∆C(t) between the two, the higher the methylation levels are, and vice versa (Figure 4). DNA visualization methods, such as an agarose gel, can also be used to determine if the digestion occurred at the region of interest.
Methylated and non-methylated DNA standards should be digested in parallel to experimental samples to demonstrate the robustness of the restriction enzyme in distinguishing methylated versus non-methylated sites. This will also help determine the sensitivity of the assay to the region of interest.
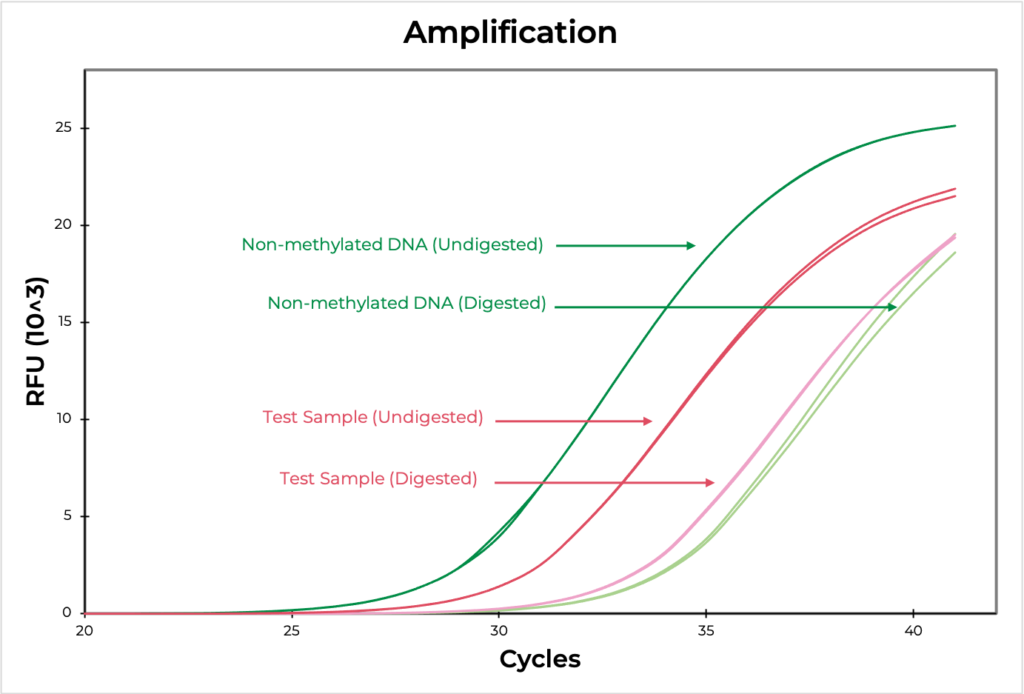
Figure 4. The amplification curve demonstrates the differences in Ct values between digested and undigested samples. The MSRE assay detected 100.9% and 2.7% methylation in the methylated and non-methylated DNA standards, respectively, confirming that the assay worked efficiently. The test sample methylation level was determined to be 17.3.
DNA STANDARDS FOR OTHER APPLICATIONS
DNA standards can be used for optimizing a variety of other applications as it is high-quality genomic DNA with known methylation patterns. This makes them ideal substitutes for precious DNA or problematic sample types when titrating experimental conditions. For example, when optimizing bisulfite library preparation of FFPE DNA, the DNA standards can easily be sonicated or fragmented to be representative of the experimental samples.
The methylated and non-methylated DNA standards have been adapted as controls for numerous methylation assays:
- #MethyLight assays 5, 6
- #Methylation-sensitive high-resolution melt analysis 7-9
- #Pyrosequencing 10, 11
- #Methylation arrays 12
- #Methylated DNA Immunoprecipitation 13
- #Bisulfite sequencing library preparation 14
The Human Methylated & Non-methylated DNA Standards are an example of a validated control set for methylation analysis of human samples. They have been widely used for all the assays mentioned above and much more. Zymo Research also provides a range of other DNA standards that can be used for optimization or quality controls for other assays to help guarantee your data is of the highest quality and ready for publication.
References:
1. Robertson, K. D., DNA methylation and human disease. Nat Rev Genet 2005, 6 (8), 597-610.
2. Roy, D.; Tiirikainen, M., Diagnostic Power of DNA Methylation Classifiers for Early Detection of Cancer. Trends Cancer 2020, 6 (2), 78-81.
3. Wick, W.; Weller, M.; van den Bent, M.; Sanson, M.; Weiler, M.; von Deimling, A.; Plass, C.; Hegi, M.; Platten, M.; Reifenberger, G., MGMT testing–the challenges for biomarker-based glioma treatment. Nat Rev Neurol 2014, 10 (7), 372-85.
4. Hernández, H. G.; Tse, M. Y.; Pang, S. C.; Arboleda, H.; Forero, D. A., Optimizing methodologies for PCR-based DNA methylation analysis. Biotechniques 2013, 55 (4), 181-97.
5. Olkhov-Mitsel, E.; Zdravic, D.; Kron, K.; van der Kwast, T.; Fleshner, N.; Bapat, B., Novel multiplex MethyLight protocol for detection of DNA methylation in patient tissues and bodily fluids. Sci Rep 2014, 4, 4432.
6. Delgado-Cruzata, L.; Vin-Raviv, N.; Tehranifar, P.; Flom, J.; Reynolds, D.; Gonzalez, K.; Santella, R. M.; Terry, M. B., Correlations in global DNA methylation measures in peripheral blood mononuclear cells and granulocytes. Epigenetics 2014, 9 (11), 1504-10.
7. Ribeiro Ferreira, I.; Darleans Dos Santos Cunha, W.; Henrique Ferreira Gomes, L.; Azevedo Cintra, H.; Lopes Cabral Guimarães Fonseca, L.; Ferreira Bastos, E.; Clinton Llerena, J.; Farias Meira de Vasconcelos, Z.; da Cunha Guida, L., A rapid and accurate methylation-sensitive high-resolution melting analysis assay for the diagnosis of Prader Willi and Angelman patients. Mol Genet Genomic Med 2019, 7 (6), e637.
8. Rajić, J.; Inic-Kanada, A.; Stein, E.; Dinić, S.; Schuerer, N.; Uskoković, A.; Ghasemian, E.; Mihailović, M.; Vidaković, M.; Grdović, N.; Barisani-Asenbauer, T., Infection Is Associated with E-Cadherin Promoter Methylation, Downregulation of E-Cadherin Expression, and Increased Expression of Fibronectin and α-SMA-Implications for Epithelial-Mesenchymal Transition. Front Cell Infect Microbiol 2017, 7, 253.
9. Rawluszko, A. A.; Bujnicka, K. E.; Horbacka, K.; Krokowicz, P.; Jagodziński, P. P., Expression and DNA methylation levels of prolyl hydroxylases PHD1, PHD2, PHD3 and asparaginyl hydroxylase FIH in colorectal cancer. BMC Cancer 2013, 13, 526.
10. Sparago, A.; Verma, A.; Patricelli, M. G.; Pignata, L.; Russo, S.; Calzari, L.; De Francesco, N.; Del Prete, R.; Palumbo, O.; Carella, M.; Mackay, D. J. G.; Rezwan, F. I.; Angelini, C.; Cerrato, F.; Cubellis, M. V.; Riccio, A., The phenotypic variations of multi-locus imprinting disturbances associated with maternal-effect variants of NLRP5 range from overt imprinting disorder to apparently healthy phenotype. Clin Epigenetics 2019, 11 (1), 190.
11. Yu, W.; Qin, X.; Jin, Y.; Li, Y.; Santiskulvong, C.; Vu, V.; Zeng, G.; Zhang, Z.; Chow, M.; Rao, J., Tianshengyuan-1 (TSY-1) regulates cellular Telomerase activity by methylation of TERT promoter. Oncotarget 2017, 8 (5), 7977-7988.
12. Van der Auwera, I.; Yu, W.; Suo, L.; Van Neste, L.; van Dam, P.; Van Marck, E. A.; Pauwels, P.; Vermeulen, P. B.; Dirix, L. Y.; Van Laere, S. J., Array-based DNA methylation profiling for breast cancer subtype discrimination. PLoS One 2010, 5 (9), e12616.
13. Rauen, T.; Grammatikos, A. P.; Hedrich, C. M.; Floege, J.; Tenbrock, K.; Ohl, K.; Kyttaris, V. C.; Tsokos, G. C., cAMP-responsive element modulator α (CREMα) contributes to decreased Notch-1 expression in T cells from patients with active systemic lupus erythematosus (SLE). J Biol Chem 2012, 287 (51), 42525-32.
14. Zheleznyakova, G. Y.; Nilsson, E. K.; Kiselev, A. V.; Maretina, M. A.; Tishchenko, L. I.; Fredriksson, R.; Baranov, V. S.; Schiöth, H. B., Methylation levels of SLC23A2 and NCOR2 genes correlate with spinal muscular atrophy severity. PLoS One 2015, 10 (3), e0121964.
Are Liquid Biopsies the Future of Diagnostics?
Are Liquid Biopsies the Future of Diagnostics?
In the constantly improving field of clinical diagnostics, a revolution is occurring in the form of liquid biopsies and cell-free nucleic acid (cfNA) analysis. Liquid biopsies are a non/less-invasive method to analyze health status from biological fluid samples (serum, saliva, urine, and other fluid sample types). Typically, they require small amounts of starting material (<10 mL), are minimally-invasive and simple to perform. Compared to liquid biopsies, surgical biopsies can be very invasive, time consuming, painful, and are limited in scope.
The significance of liquid biopsies lies in their ability to detect early disease with just a simple blood draw. As technologies improve the detection of specific disease biomarkers, earlier adoption of more effective targeted treatments is possible. This potential has been gaining a lot of attention, leading to significant investments by companies like Illumina and Roche.
Cell-free Nucleic Acids
It has long been recognized that biological fluids contain DNA and RNA that is not contained in cells. It is now known that these cell-free nucleic acids are released into biological fluids from apoptotic and necrotic cells or secreted from healthy and diseased cells via microvesicles such as exosomes [1]. The presence of cfNA in biological liquids has been long recognized, but its function is poorly understood, as practical uses have been hampered by insufficient isolation and analytical methods [2]. For example, cfNAs have been linked to cancer since their discovery in 1948 [3], but due to technical limitations, it took almost 20 years for a link between cell-free DNA (cfDNA) and cancer to be established and widely supported.
It took another 20 years to define tumor-derived cfDNA within samples taken from cancer patients [4]. However, with the inception of the Human Genome Project and recent technological advances in genome analysis, the field of cell-free nucleic acid research has grown rapidly and cell-free RNA (cfRNA) analysis is starting to gain recognition as a source of important biomarkers [5].
The use of cell-free nucleic acid detection has expanded beyond cancer detection. Liquid biopsy methods are now being evaluated for diagnostic effectiveness in disease and pre-natal genetic testing. If proven effective, these techniques will allow for safer and earlier in vivo detection of various inherited genetic diseases. The goal is to implement personalized medical interventions in early stages of disease for a better chance of positive effects on health outcomes. Many tests already widely used with cell-free fetal DNA (cffDNA) include a first trimester Down syndrome screen, amniotic fluid analysis, and chromosomal karyotype analysis.
Many applications are currently being evaluated as a less-invasive method to monitor patient health. These include testing for fetal aneuploidy, monitoring the status of organ transplants, blunt trauma and burn recovery progression, and detection of sepsis and septic shock in patients [6]. In addition to these, research is underway to use cfNA for early diagnosis of genetic disorders such as β-thalassemia and sickle cell anemia [6]. Earlier detection will lead to earlier treatment of many of these disorders, which will improve treatment outcomes and prevent comorbidities from developing.
Sample Diversity and Purification Issues
Numerous biofluids such as blood, cerebrospinal fluid (CSF), saliva, amniotic fluid, and urine [4, 6-9] contain readily detectable cfNA populations. However, each of these sample sources produce specific challenges during collection, handling, and extraction that must be addressed to obtain accurate profiles. Cell-free DNA is most commonly isolated from the serum or plasma fraction of blood, however extraction methods must also be able to address a wide range of sample types.
The most readily available methods to collect cfDNA have been restricted to commercially available extraction kits or homemade methods. These methods may not be optimized and often use outdated technologies. Cell-free biofluids are challenging to work with due to their high protein content and low amount of cfNA in relatively large amounts of liquid. This is why traditional cfNA extraction technologies have been so limited in isolation efficiency, sample compatibility, and have been unable to isolate both cfDNA and cfRNA from the same input sample.
To address this issue, Zymo Research produced a revolutionary kit designed to easily process samples from liquid biopsies, all in one quick, simple workflow. The Quick-cfDNA/cfRNA™ Serum & Plasma Kit contains protocols that make it easy to isolate cfDNA from even the most difficult sample types (Figure 1).
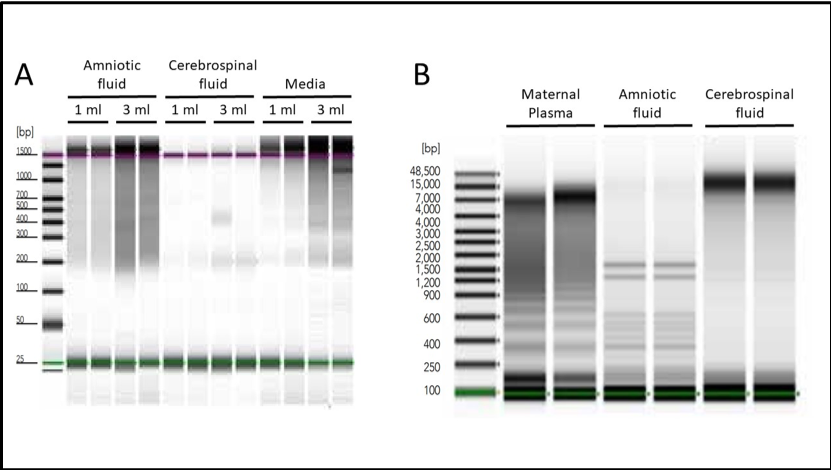
Figure 1: Isolation of cell-free nucleic acids from various sources. A. Amniotic fluid, cerebrospinal fluid, or spent HeLa cell culture media were used to isolate cfNAs using Quick-cfDNA/cfRNA Serum & Plasma Kit. Endogenous cell-free DNA from each sample type was visualized using the Agilent 2200 TapeStation® system. B. Total cfDNA, including both high and low molecular weight species, was purified in duplicate from human maternal plasma, amniotic fluid, and cerebrospinal fluid using the Quick-cfDNA Serum & Plasma kit. DNA was visualized using the Agilent 2200 TapeStation® system.
Cell-free Nucleic Acid Profile Analysi
Another challenge facing cfNA analysis is the robustness of samples when initiated into sequencing pipelines such as RNA sequencing (RNA-Seq). Along with the relatively low amount of nucleic acid, microRNAs are often lost during purification which leads to profile bias. This is can be due to the use of non-optimal extraction methods that produce biased cfNA recovery. For example, three commercial methods of cfRNA extraction from plasma demonstrates difference in the cell-free microRNA yield for hsa-miR-16-5p in the figure below (Figure 2).
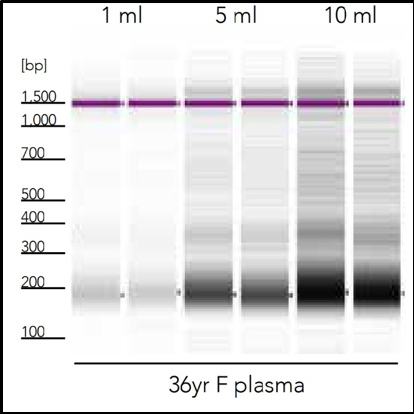
Figure 2: Cell-free DNA recovery scales proportionally with sample input. Cell-free DNA was isolated in duplicate from three healthy female donors using the Quick-cfDNA Serum & Plasma kit and visualized using the Agilent 2200 TapeStation® system.
Cell-free Nucleic Acid Profile Analysis
Another challenge facing cfNA analysis is the robustness of samples when initiated into sequencing pipelines such as RNA sequencing (RNA-Seq). Along with the relatively low amount of nucleic acid, microRNAs are often lost during purification which leads to profile bias. This is can be due to the use of non-optimal extraction methods that produce biased cfNA recovery. For example, three commercial methods of cfRNA extraction from plasma demonstrates difference in the cell-free microRNA yield for hsa-miR-16-5p in the figure below (Figure 3).
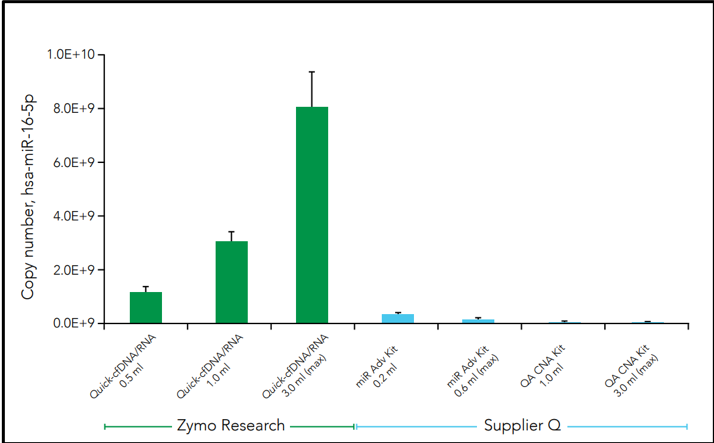
Figure 3: Isolation of human miR-16-5p using commercially available cfNA extraction kits. Samples from the same donor (plasma from 61y-F) were processed using the manufacturers’ suggested protocol and eluted in 30 µl. Quantification of human hsa-miR-16-5p was assayed using the method described by Busk [10]. The yield scales linearly to input volume for Quick-cfDNA/cfRNA Serum & Plasma Kit.
Delicate Yet Difficult
While the information provided by this RNA/DNA ratio is critical to understanding the risks posed by environmental stress, purifying DNA and RNA from coral sample can be tricky. Coral tissues contain strong collagen fibers, which can make cells lysis difficult. However, high-quality DNA and RNA can still be isolated and purified from coral tissue with special considerations. For example, it is suggested to remove as much skeleton as possible from the coral tissue in order to minimize the possible effects of endolithic organisms.
Below are a few recommended protocols for processing coral samples.
Further comparative analysis of commercial kit cfRNA extracts followed by RNA-seq analysis demonstrates that the Quick-cfRNA Serum & Plasma kit yielded higher quality reads, better alignment to sequence databases, and achieved recovery of 463 additional microRNA species not recovered by the supplier Q kit (Table 1).

Table 1: Comparison of RNA-Seq analysis of cell-free microRNAs from two commercial kit workflows. Total cell-free RNAs were isolated from 200 µl plasma from each of four different donors. RealSeq-Biofluids Library Prep Kit (SomaGenics) was used to generate RNA sequencing library and ran on MiniSeq System (Illumina). Read averages, quality, alignments, and microRNA diversity quantification are summarized above.
Conclusion
The field of liquid biopsy is rapidly growing due to the massive technological revolution in genomics over the past 30 years. The discovery of cell-free nucleic acids has allowed for doctors to monitor cancer and other disease states in human adults as well as detect complications early on in pregnancy. The advent of analytical methods for cell-free nucleic acid detection has allowed for significantly safer disease monitoring and pregnancy testing. This is because rather than a surgical biopsy or amniocentesis physicians can get all the information from a simple blood draw. With these tools in place, scientists and doctors can work together to bring us to a healthier and safer future through early diagnosis and rapid disease monitoring.
References:
[1] Dominguez-Vigil IG, Moreno-Martinez AK, Wang JY, Roehrl MHA, Barrera-Saldana HA. The dawn of the liquid biopsy in the fight against cancer. Oncotarget. 2018;9:2912-22.
[2] Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24.
[3] Mandel P, Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil. 1948;142:241-3.
[4] Bryzgunova OE, Laktionov PP. Extracellular Nucleic Acids in Urine: Sources, Structure, Diagnostic Potential. Acta Naturae. 2015;7:48-54.
[5] Zaporozhchenko IA, Ponomaryova AA, Rykova EY, Laktionov PP. The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn. 2018;18:133-45.
[6] Soltani M, Nemati M, Maralani M, Estiar MA, Andalib S, Fardiazar Z, et al. Cell-free fetal DNA in amniotic fluid supernatant for prenatal diagnosis. Cell Mol Biol (Noisy-le-grand). 2016;62:14-7.
[7] Connolly ID, Li Y, Pan W, Johnson E, You L, Vogel H, et al. A pilot study on the use of cerebrospinal fluid cell-free DNA in intramedullary spinal ependymoma. J Neurooncol. 2017;135:29-36.
[8] Hui L, Bianchi DW. Cell-free fetal nucleic acids in amniotic fluid. Hum Reprod Update. 2011;17:362-71.
[9] Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17:3.
[10] Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics. 2014;15:29.

