 |
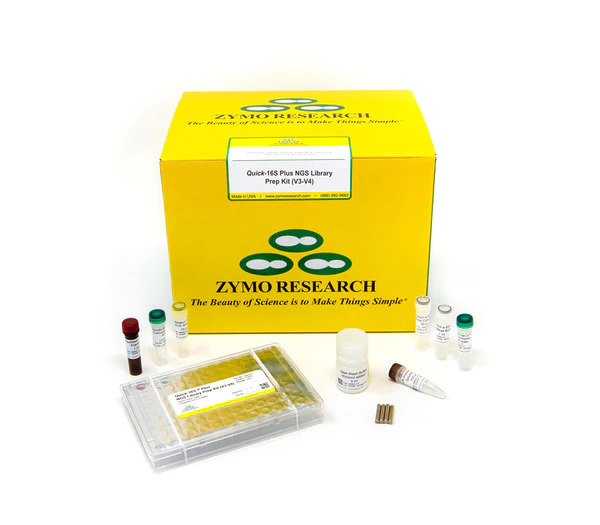
Quick-16S Plus NGS Library Prep Kit (V3-V4, UDI)
The Quick-16S Plus NGS Library Prep Kit (V3-V4, UDI) is the fastest and simplest NGS library prep targeting the V3-V4 region of the 16S rRNA gene for high-throughput sequencing. |
 |
ZymoPURE 96 Plasmid Miniprep Kit
The ZymoPURE™ 96 Plasmid Miniprep Kit features a high-throughput method for the purification of up to 100 µg of ultra-pure transfection-grade plasmid DNA using a vacuum manifold or centrifuge |
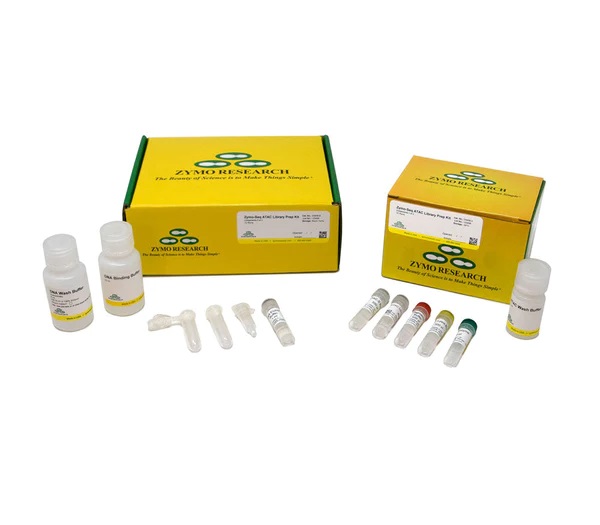 |
Zymo-Seq ATAC Library Kit
ymo-Seq ATAC Library Kit provides a simple and streamlined workflow for Assay for Transposase Accessible Chromatin with high throughput Sequencing (ATAC-Seq) library preparation from mammalian cell and tissue input. This all-inclusive kit features an easy to follow protocol capable of preparing ATAC-Seq libraries from as little as 50,000 cells in as little as 4 hours with minimal hands on time. |
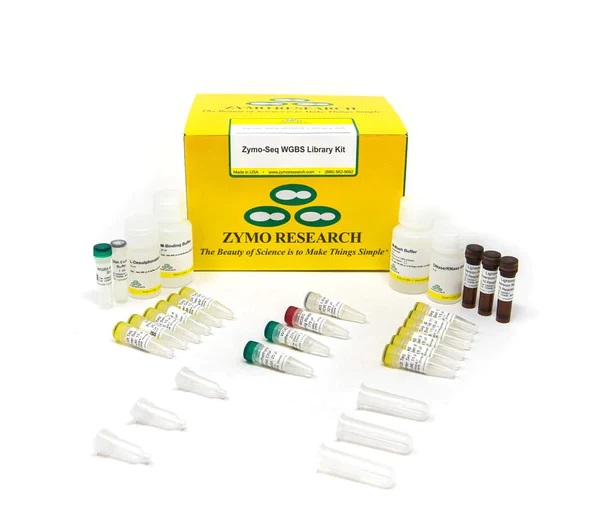 |
Zymo-Seq WGBS Library Kit
Zymo-Seq WGBS Library Kit is the only kit available for whole genome bisulfite (WGBS) library preparation in a single tube. Zymo-Seq WGBS Library Kit incorporates tagmentation technology to eliminate the tedious fragmentation, enzymatic, and clean-up steps required by conventional, ... |
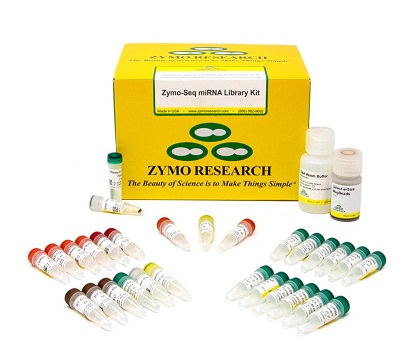 |
Zymo-Seq miRNA Library Kit
The Zymo-Seq™ miRNA Library Kit is an innovative small RNA sequencing library prep kit that uses a single-adapter and circularization strategy to reduce ligation bias and provide accurate small RNA profiling from cell-free RNA (cfRNA) and total RNA. This simple, streamlined workflow minimizes hands-on time and uses gel-free removal of adapter dimers to generate stranded small RNA libraries from plasma, biofluids, cells, tissues, and more. |
 |
ZymoBIOMICS DNA & RNA Kits
UNBIASED DNA AND RNA EXTRACTION FOR MICROBIOME PROFILING |
 |
ZymoBIOMICS 96 MagBead DNA Kit
ZymoBIOMICS 96 MagBead DNA Kit |
 |
Zymo-Seq Cell Free DNA WGBS Library Kit
Zymo-Seq Cell Free DNA WGBS Library Kit |
 |
DNA Clean & Concentrator MagBead Kit
The DNA Clean & Concentrator MagBead Kit is magnetic bead based clean-up kit that can purify DNA from PCR, enzymatic reactions, impure extractions, and other sources. The MagBinding Bead technology provides a rapid and scalable workflow that can be adapted for low to high-throughput automated method for purification and concentration of DNA. |
 |
Quick-16S Plus NGS Library Prep Kit (V4)
The Quick-16S Plus NGS Library Prep Kit (V4) is the fastest and simplest NGS library prep targeting the V4 region of the 16S rRNA gene for high-throughput sequencing. The automation-friendly protocol utilizes a premixed amplification plate and a single qPCR/PCR for combined targeted amplification and barcode addition. Pooling by equal volume and a single clean-up of the final library, rather than massive AMPure® bead-based clean-ups, simplifies the library prep process. Additional library quantification analysis such as TapeStation® analysis or gel electrophoresis are not necessary. With these features, the workflow dramatically reduces the hands-on time of library preparation to only 30 minutes. |


 We're here to help
We're here to help  High Quality Guaranteed Product
High Quality Guaranteed Product