New SARS-CoV-2 Recombinant Virus Lineages
Three SARS-CoV-2 recombinant lineages have been designated by Pangolin as of March 2022, two a combination of Delta and BA.1 (designated XD and XF), and the third a combination of Omicron BA.1 and Omicron BA.2 with a few additional mutations in NSP3 and NSP12 (designated XE).
Below is a graphical representation of the recombinant lineages as compared to the genome schematic, courtesy of the UK Health Security Agency.
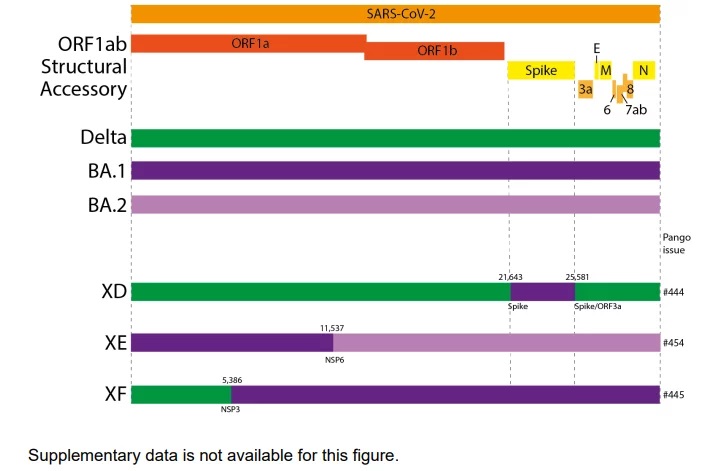
SARS-CoV-2 XD, XE, and XF Epidemiology
As mentioned, XD is a combination of Delta and Omicron BA.1. More specifically, it is comprised of Delta AY.4 with an Omicron BA.1 Spike. Recombination of two different strains combines features of both parent viruses, potentially causing different phenotypic expressions than either parent virus. In this case potentially the infectivity of Omicron and the disease severity of Delta. The earliest samples were collected in December 2021 and to date, samples have been identified in France, Denmark, and Belgium.
XE, however, is a combination of two Omicron sub-strains, BA.1 and BA.2, first detected on January 19, 2022. This variant contains NSP1-6 from BA.1 with the remainder of the genome from BA.2 with the recombination occurring at nucleotide 11,537. According to the WHO, it currently has an estimated community growth rate advantage of 1.1 as compared to BA.2, which represents a 10% increase in transmissibility.
XF, while also recombination of Delta and Omicron BA.1, is instead comprised of the first part of the genome up until nucleotide 5386, near the end of NSP3. This variant appears to be unique to the UK with samples found between January 7, 2022, and February 14, 2022, and no current evidence of growth.
While it is too early to tell if these recombinant variants will cause a public health concern, it is to be expected as viruses recombine all the time.
Viral Recombination
Recombination occurs in many viruses, including other coronaviruses, HIV, and influenza. Results of recombination vary from being innocuous to potentially generate more infectious virions.
How do viruses recombine? This phenomenon occurs when two strains of virus infect the same host cells and interact to generate another strain during replication that contains genes from both original strains.
RNA viruses undergo two forms of recombination. One is copy choice recombination, which occurs during viral replication when the RNA polymerase switches from one RNA to another, creating a new RNA strand that is a combination of the two. The other is reassortment, occurring in viruses with segmented genomes, where different segments of different genomes are packaged into a new virion.
Further your Research with ProSci
To date, ProSci has developed variant-specific reagents by focusing on unique point mutations in the spike protein. Cat. No. 9793 targets the G142D Δ143-145VYY mutation and deletion combination found in BA.1 and Cat. No. 9805 targets the Q493R G496S Q498R N501Y Y505H region currently unique to BA.1 (4 mutations shared with BA.2).
To further research, recombinant proteins for BA.1 (RBD Cat. No. 95-128 and Spike S1 Cat. No. 95-129) are available as well as a BA.1 variant pseudovirus (Cat. No. 95-201). For additional variant-specific research reagents and wild-type controls, visit our Variants Research Reagents.
At the onset of the COVID-19 pandemic, ProSci developed antibodies against SARS-CoV-2 and has continued to expand its catalog of antibodies and recombinant proteins. Explore the comprehensive catalog of ProSci SARS-CoV-2 Research Reagents.
References:
- New SARS-CoV-2 Recombinant Virus Lineages | ProSci Incorporated (prosci-inc.com)
- UK Health Security Agency, Technical briefing 39, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1063424/Tech-Briefing-39-25March2022_FINAL.pdf
- https://twitter.com/PeacockFlu/status/1504158922675998723
- Weekly epidemiological update on COVID-19 – 5 April 2022, Edition 86. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- Fleischmann, Robert. Medical Microbiology. Galveston, TX, Univ of Texas Medical Branch, 1996. https://www.ncbi.nlm.nih.gov/books/NBK8439/
- Simon-Loriere, E., Holmes, E. Why do RNA viruses recombine? Nat Rev Microbiol9, 617–626 (2011). https://doi.org/10.1038/nrmicro2614

